Enzymatic attributes of an l-isoaspartyl methyltransferase from Candida utilis and its role in cell survival
- PMID: 29124188
- PMCID: PMC5668901
- DOI: 10.1016/j.bbrep.2015.08.015
Enzymatic attributes of an l-isoaspartyl methyltransferase from Candida utilis and its role in cell survival
Abstract
Backgrounds: Spontaneous deamidation and isoaspartate (IsoAsp) formation contributes to aging and reduced longevity in cells. A protein-l-isoaspartate (d-aspartate) O-methyltransferase (PCMT) is responsible for minimizing IsoAsp moieties in most organisms.
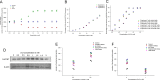
Methods: PCMT was purified in its native form from yeast Candida utilis. The role of the native PCMT in cell survival and protein repair was investigated by manipulating intracellular PCMT levels with Oxidized Adenosine (AdOx) and Lithium Chloride (LiCl). Proteomic Identification of possible cellular targets was carried out using 2-dimensional gel electrophoresis, followed by on-Blot methylation and mass spectrometric analysis.
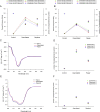
Results: The 25.4 kDa native PCMT from C. utilis was found to have a Km of 3.5 µM for AdoMet and 33.36 µM for IsoAsp containing Delta Sleep Inducing Peptide (DSIP) at pH 7.0. Native PCMT comprises of 232 amino acids which is coded by a 698 bp long nucleotide sequence. Phylogenetic comparison revealed the PCMT to be related more closely with the prokaryotic homologs. Increase in PCMT levels in vivo correlated with increased cell survival under physiological stresses. PCMT expression was seen to be linked with increased intracellular reactive oxygen species (ROS) concentration. Proteomic identification of possible cellular substrates revealed that PCMT interacts with proteins mainly involved with cellular housekeeping. PCMT effected both functional and structural repair in aged proteins in vitro.
General significance: Identification of PCMT in unicellular eukaryotes like C. utilis promises to make investigations into its control machinery easier owing to the familiarity and flexibility of the system.
Keywords: Deamidation; Enzyme catalysis; Enzyme purification; Isoaspartate; MALDI TOF; S-adenosyl l-methionine; Yeast.
Figures



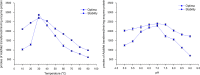

 ), pH (
), pH ( ), hyperosmotic (
), hyperosmotic ( ) and heat (
) and heat ( ) stress. Isoapartate levels of treated and untreated cells were worked out in terms of pmoles of isoAsp/pmole total cellular proteins. (F) Early stationary phase Candida cells were incubated in presence or absence of 1.2 mM LiCl for 24 h after which the cells were subjected to either oxidative (
) stress. Isoapartate levels of treated and untreated cells were worked out in terms of pmoles of isoAsp/pmole total cellular proteins. (F) Early stationary phase Candida cells were incubated in presence or absence of 1.2 mM LiCl for 24 h after which the cells were subjected to either oxidative ( ), pH (
), pH ( ), hyperosmotic (
), hyperosmotic ( ) and heat (
) and heat ( ) stress. Intracellular isoAsp levels from control and treated sets were determined in terms of pmoles of isoAsp/pmole total cellular proteins. Data presented is from two independent experimentation sets. The horizontal bars indicate the average (Mean), and the error bars represent standard deviation (S.D.).
) stress. Intracellular isoAsp levels from control and treated sets were determined in terms of pmoles of isoAsp/pmole total cellular proteins. Data presented is from two independent experimentation sets. The horizontal bars indicate the average (Mean), and the error bars represent standard deviation (S.D.).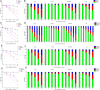

Similar articles
-
Protein isoaspartate methyltransferase prevents apoptosis induced by oxidative stress in endothelial cells: role of Bcl-Xl deamidation and methylation.PLoS One. 2008 Sep 22;3(9):e3258. doi: 10.1371/journal.pone.0003258. PLoS One. 2008. Retraction in: PLoS One. 2018 Nov 8;13(11):e0207530. doi: 10.1371/journal.pone.0207530. PMID: 18806875 Free PMC article. Retracted.
-
Structural integrity of histone H2B in vivo requires the activity of protein L-isoaspartate O-methyltransferase, a putative protein repair enzyme.J Biol Chem. 2001 Oct 5;276(40):37161-5. doi: 10.1074/jbc.M106682200. Epub 2001 Jul 30. J Biol Chem. 2001. PMID: 11479322
-
Immunochemical characterization of L-isoaspartyl-protein carboxyl methyltransferase from mammalian tissues.Biochem J. 1995 Aug 1;309 ( Pt 3)(Pt 3):993-8. doi: 10.1042/bj3090993. Biochem J. 1995. PMID: 7639720 Free PMC article.
-
PIMT-Mediated Protein Repair: Mechanism and Implications.Biochemistry (Mosc). 2019 May;84(5):453-463. doi: 10.1134/S0006297919050018. Biochemistry (Mosc). 2019. PMID: 31234761 Review.
-
PROTEIN l-ISOASPARTYL METHYLTRANSFERASE (PIMT) in plants: regulations and functions.Biochem J. 2020 Nov 27;477(22):4453-4471. doi: 10.1042/BCJ20200794. Biochem J. 2020. PMID: 33245750 Review.
Cited by
-
Nitrogen Supply and Host-Plant Genotype Modulate the Transcriptomic Profile of Plasmodiophora brassicae.Front Microbiol. 2021 Jul 8;12:701067. doi: 10.3389/fmicb.2021.701067. eCollection 2021. Front Microbiol. 2021. PMID: 34305867 Free PMC article.
References
-
- Clarke S. World Scientific Publishings; NJ: 1999. A protein carboxyl methyltransfease that recognizes and repairs age-damaged peptides and proteins and participates in their repair.
-
- Aswad D.W., Paranandi M.V., Schurter B.T. Isoaspartate in peptides and proteins: formation, significance, and analysis. J. Pharm. Biomed. Anal. 2000;21:1129–1136. - PubMed
-
- Lowenson J.D., Clarke S. Structural elements affecting the recognition of l-isoaspartyl residues by the l-isoaspartyl/d-aspartyl protein methyltransferase. Implications for the repair hypothesis. J. Biol. Chem. 1991;266:19396–19406. - PubMed
-
- Furuchi T., Sakurako K., Katane M., Sekine M., Homma H. The role of protein l-Isoaspartyl/d-aspartyl O-methyltransferase (PIMT) in intracellular signal transduction. Chem Biodivers. 2010;7:1337–1348. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

