Biophysical and enzymatic properties of aminoglycoside adenylyltransferase AadA6 from Pseudomonas aeruginosa
- PMID: 29124199
- PMCID: PMC5668923
- DOI: 10.1016/j.bbrep.2015.09.011
Biophysical and enzymatic properties of aminoglycoside adenylyltransferase AadA6 from Pseudomonas aeruginosa
Abstract
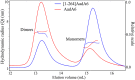
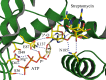
The gene coding for the aminoglycoside adenylyltransferase (aadA6) from a clinical isolate of Pseudomonas aeruginosa was cloned and expressed in Escherichia coli strain BL21(DE3)pLysS. The overexpressed enzyme (AadA6, 281 amino-acid residues) and a carboxy-terminal truncated variant molecule ([1-264]AadA6) were purified to near homogeneity and characterized. Light scattering experiments conducted under low ionic strength supported equilibrium between monomeric and homodimeric arrangements of the enzyme subunits. Circular Dichroism spectropolarimetry indicated a close structural relation to adenylate kinases. Both forms modified covalently the aminoglycosides streptomycin and spectinomycin. The enzyme required at least 5 mM MgCl2 for normal Michaelis-Menten kinetics. Streptomycin exhibited a strong substrate inhibition effect at 1 mM MgCl2. The truncated 17 residues at the C-terminus have little influence on protein folding, whereas they have a positive effect on the enzymic activity and stabilize dimers at high protein concentrations (>100 μM). Homology modelling and docking based on known crystal structures yielded models of the central ternary complex of monomeric AadA6 with ATP and streptomycin or spectinomycin.
Keywords: AMPCPP, α,β-methyleneadenosine 5′-triphosphate; Aminoglycoside adenylyltransferase; Antibiotic modification; Circular dichroism; Enzyme kinetics; Homology modelling; MALS, Multi-angle light scattering; MIC, Minimum Inhibitory Concentration; Multi-angle light scattering; [1-264]AadA6, genetically truncated AadA6 at the carboxy-terminus; [1-281]AadA6 or AadA6, full length aminoglycoside adenylyltransferase.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

