Sequencing and De Novo Assembly of the Toxicodendron radicans (Poison Ivy) Transcriptome
- PMID: 29125533
- PMCID: PMC5704230
- DOI: 10.3390/genes8110317
Sequencing and De Novo Assembly of the Toxicodendron radicans (Poison Ivy) Transcriptome
Abstract
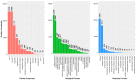
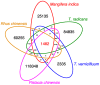
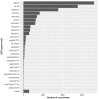
Contact with poison ivy plants is widely dreaded because they produce a natural product called urushiol that is responsible for allergenic contact delayed-dermatitis symptoms lasting for weeks. For this reason, the catchphrase most associated with poison ivy is "leaves of three, let it be", which serves the purpose of both identification and an appeal for avoidance. Ironically, despite this notoriety, there is a dearth of specific knowledge about nearly all other aspects of poison ivy physiology and ecology. As a means of gaining a more molecular-oriented understanding of poison ivy physiology and ecology, Next Generation DNA sequencing technology was used to develop poison ivy root and leaf RNA-seq transcriptome resources. De novo assembled transcriptomes were analyzed to generate a core set of high quality expressed transcripts present in poison ivy tissue. The predicted protein sequences were evaluated for similarity to SwissProt homologs and InterProScan domains, as well as assigned both GO terms and KEGG annotations. Over 23,000 simple sequence repeats were identified in the transcriptome, and corresponding oligo nucleotide primer pairs were designed. A pan-transcriptome analysis of existing Anacardiaceae transcriptomes revealed conserved and unique transcripts among these species.
Keywords: Anacardiaceae; Toxicodendron radicans; contact dermatitis; poison ivy; skin rash; transcriptome; urushiol.
Conflict of interest statement
The authors declare no conflict of interest. The Virginia Bioinformatics Institute Core Lab (funding sponsor) performed the Illumina DNA libraries and performed the Illumina DNA sequencing. With the notable exception of the collection of the Illumina sequence data, the founding sponsors had no role in the design of the study; in the analyses, or interpretation of the data; in the writing of the manuscript, and in the decision to publish the results.
Figures
Similar articles
-
Toxicodendron Toxicity.2023 May 16. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 16. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 32491789 Free Books & Documents.
-
Characterization of 42 microsatellite markers from poison ivy, Toxicodendron radicans (Anacardiaceae).Int J Mol Sci. 2013 Oct 14;14(10):20414-26. doi: 10.3390/ijms141020414. Int J Mol Sci. 2013. PMID: 24129176 Free PMC article.
-
The complete genome sequence of Toxicodendron radicans, Eastern Poison Ivy.F1000Res. 2020 Aug 20;9:1015. doi: 10.12688/f1000research.25556.1. eCollection 2020. F1000Res. 2020. PMID: 34249347 Free PMC article.
-
Toxicodendron dermatitis: poison ivy, oak, and sumac.Wilderness Environ Med. 2006 Summer;17(2):120-8. doi: 10.1580/pr31-05.1. Wilderness Environ Med. 2006. PMID: 16805148 Review.
-
Occupational poison ivy and oak dermatitis.Dermatol Clin. 1994 Jul;12(3):511-6. Dermatol Clin. 1994. PMID: 7923948 Review.
Cited by
-
Reference nodule transcriptomes for Melilotus officinalis and Medicago sativa cv. Algonquin.Plant Direct. 2022 Jun 8;6(6):e408. doi: 10.1002/pld3.408. eCollection 2022 Jun. Plant Direct. 2022. PMID: 35774624 Free PMC article.
-
Pilot RNA-seq data from 24 species of vascular plants at Harvard Forest.Appl Plant Sci. 2021 Feb 14;9(2):e11409. doi: 10.1002/aps3.11409. eCollection 2021 Feb. Appl Plant Sci. 2021. PMID: 33680580 Free PMC article.
-
Transcriptome Profiling Provides Insight into the Genes in Carotenoid Biosynthesis during the Mesocarp and Seed Developmental Stages of Avocado (Persea americana).Int J Mol Sci. 2019 Aug 23;20(17):4117. doi: 10.3390/ijms20174117. Int J Mol Sci. 2019. PMID: 31450745 Free PMC article.
-
The leaf transcriptome of fennel (Foeniculum vulgare Mill.) enables characterization of the t-anethole pathway and the discovery of microsatellites and single-nucleotide variants.Sci Rep. 2018 Jul 11;8(1):10459. doi: 10.1038/s41598-018-28775-2. Sci Rep. 2018. PMID: 29993007 Free PMC article.
-
The genome evolution and domestication of tropical fruit mango.Genome Biol. 2020 Mar 6;21(1):60. doi: 10.1186/s13059-020-01959-8. Genome Biol. 2020. PMID: 32143734 Free PMC article.
References
-
- Epstein W.L. Occupational poison ivy and oak dermatitis. Derrmatol. Clin. 1994;3:511–516. - PubMed
-
- Gayer K.D., Burnett J.W. Toxicodendron dermatitis. Cutis. 1988;42:99–100. - PubMed
-
- Mitchell J., Rook A. Botanical Dermatology: Plants and Plant Products Injurious to the Skin. Greengrass; Vancouver, BC, Canada: 1979. p. 787.
-
- Hill G.A., Mattacotti V., Graha W.D. The toxic principle of the poison ivy. J. Am. Chem. Soc. 1934;56:2736–2738. doi: 10.1021/ja01327a064. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

