Genomics-based non-invasive prenatal testing for detection of fetal chromosomal aneuploidy in pregnant women
- PMID: 29125628
- PMCID: PMC6486016
- DOI: 10.1002/14651858.CD011767.pub2
Genomics-based non-invasive prenatal testing for detection of fetal chromosomal aneuploidy in pregnant women
Abstract
Background: Common fetal aneuploidies include Down syndrome (trisomy 21 or T21), Edward syndrome (trisomy 18 or T18), Patau syndrome (trisomy 13 or T13), Turner syndrome (45,X), Klinefelter syndrome (47,XXY), Triple X syndrome (47,XXX) and 47,XYY syndrome (47,XYY). Prenatal screening for fetal aneuploidies is standard care in many countries, but current biochemical and ultrasound tests have high false negative and false positive rates. The discovery of fetal circulating cell-free DNA (ccfDNA) in maternal blood offers the potential for genomics-based non-invasive prenatal testing (gNIPT) as a more accurate screening method. Two approaches used for gNIPT are massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS).
Objectives: To evaluate and compare the diagnostic accuracy of MPSS and TMPS for gNIPT as a first-tier test in unselected populations of pregnant women undergoing aneuploidy screening or as a second-tier test in pregnant women considered to be high risk after first-tier screening for common fetal aneuploidies. The gNIPT results were confirmed by a reference standard such as fetal karyotype or neonatal clinical examination.
Search methods: We searched 13 databases (including MEDLINE, Embase and Web of Science) from 1 January 2007 to 12 July 2016 without any language, search filter or publication type restrictions. We also screened reference lists of relevant full-text articles, websites of private prenatal diagnosis companies and conference abstracts.
Selection criteria: Studies could include pregnant women of any age, ethnicity and gestational age with singleton or multifetal pregnancy. The women must have had a screening test for fetal aneuploidy by MPSS or TMPS and a reference standard such as fetal karyotype or medical records from birth.
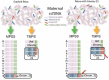
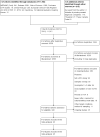
Data collection and analysis: Two review authors independently carried out study selection, data extraction and quality assessment (using the QUADAS-2 tool). Where possible, hierarchical models or simpler alternatives were used for meta-analysis.
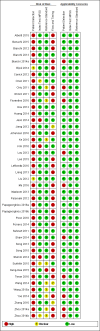
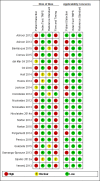
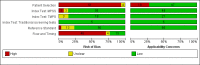
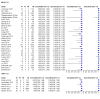
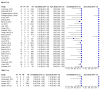
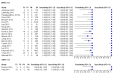
Main results: Sixty-five studies of 86,139 pregnant women (3141 aneuploids and 82,998 euploids) were included. No study was judged to be at low risk of bias across the four domains of the QUADAS-2 tool but applicability concerns were generally low. Of the 65 studies, 42 enrolled pregnant women at high risk, five recruited an unselected population and 18 recruited cohorts with a mix of prior risk of fetal aneuploidy. Among the 65 studies, 44 evaluated MPSS and 21 evaluated TMPS; of these, five studies also compared gNIPT with a traditional screening test (biochemical, ultrasound or both). Forty-six out of 65 studies (71%) reported gNIPT assay failure rate, which ranged between 0% and 25% for MPSS, and between 0.8% and 7.5% for TMPS.In the population of unselected pregnant women, MPSS was evaluated by only one study; the study assessed T21, T18 and T13. TMPS was assessed for T21 in four studies involving unselected cohorts; three of the studies also assessed T18 and 13. In pooled analyses (88 T21 cases, 22 T18 cases, eight T13 cases and 20,649 unaffected pregnancies (non T21, T18 and T13)), the clinical sensitivity (95% confidence interval (CI)) of TMPS was 99.2% (78.2% to 100%), 90.9% (70.0% to 97.7%) and 65.1% (9.16% to 97.2%) for T21, T18 and T13, respectively. The corresponding clinical specificity was above 99.9% for T21, T18 and T13.In high-risk populations, MPSS was assessed for T21, T18, T13 and 45,X in 30, 28, 20 and 12 studies, respectively. In pooled analyses (1048 T21 cases, 332 T18 cases, 128 T13 cases and 15,797 unaffected pregnancies), the clinical sensitivity (95% confidence interval (CI)) of MPSS was 99.7% (98.0% to 100%), 97.8% (92.5% to 99.4%), 95.8% (86.1% to 98.9%) and 91.7% (78.3% to 97.1%) for T21, T18, T13 and 45,X, respectively. The corresponding clinical specificities (95% CI) were 99.9% (99.8% to 100%), 99.9% (99.8% to 100%), 99.8% (99.8% to 99.9%) and 99.6% (98.9% to 99.8%). In this risk group, TMPS was assessed for T21, T18, T13 and 45,X in six, five, two and four studies. In pooled analyses (246 T21 cases, 112 T18 cases, 20 T13 cases and 4282 unaffected pregnancies), the clinical sensitivity (95% CI) of TMPS was 99.2% (96.8% to 99.8%), 98.2% (93.1% to 99.6%), 100% (83.9% to 100%) and 92.4% (84.1% to 96.5%) for T21, T18, T13 and 45,X respectively. The clinical specificities were above 100% for T21, T18 and T13 and 99.8% (98.3% to 100%) for 45,X. Indirect comparisons of MPSS and TMPS for T21, T18 and 45,X showed no statistical difference in clinical sensitivity, clinical specificity or both. Due to limited data, comparative meta-analysis of MPSS and TMPS was not possible for T13.We were unable to perform meta-analyses of gNIPT for 47,XXX, 47,XXY and 47,XYY because there were very few or no studies in one or more risk groups.
Authors' conclusions: These results show that MPSS and TMPS perform similarly in terms of clinical sensitivity and specificity for the detection of fetal T31, T18, T13 and sex chromosome aneuploidy (SCA). However, no study compared the two approaches head-to-head in the same cohort of patients. The accuracy of gNIPT as a prenatal screening test has been mainly evaluated as a second-tier screening test to identify pregnancies at very low risk of fetal aneuploidies (T21, T18 and T13), thus avoiding invasive procedures. Genomics-based non-invasive prenatal testing methods appear to be sensitive and highly specific for detection of fetal trisomies 21, 18 and 13 in high-risk populations. There is paucity of data on the accuracy of gNIPT as a first-tier aneuploidy screening test in a population of unselected pregnant women. With respect to the replacement of invasive tests, the performance of gNIPT observed in this review is not sufficient to replace current invasive diagnostic tests.We conclude that given the current data on the performance of gNIPT, invasive fetal karyotyping is still the required diagnostic approach to confirm the presence of a chromosomal abnormality prior to making irreversible decisions relative to the pregnancy outcome. However, most of the gNIPT studies were prone to bias, especially in terms of the selection of participants.
Conflict of interest statement
Mylene Badeau: none known.
Jonatan Blais: none known.
Yves Giguère (and some other members of the review team ‐ FR, SL, FL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors may eventually publish results from clinical trials that could be considered in this review. None of the four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest
Sylvie Langlois (and some other members of the review team ‐ FR, YG, FL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors (FR, SL, YG and FL) may eventually publish results from clinical trials that could be considered in this review. None of these four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest. This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
France Légaré (and some other members of the review team ‐ FR, YG, SL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors (FR, SL, YG and FL) may eventually publish results from clinical trials that could be considered in this review. None of these four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest. This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
Carmen Lindsay: none known.
Leon Nshimyumukiza: none known.
François Rousseau (and some other members of the review team ‐ SL, YG, FL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors (FR, SL, YG and FL) may eventually publish results from clinical trials that could be considered in this review. None of these four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest. This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
Yemisi Takwoingi: The University of Birmingham received consultancy fees from Universite Laval for statistical analysis.
Alexis F Turgeon: This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
William Witteman: none known.
Figures


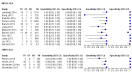


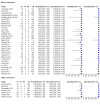
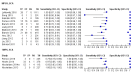

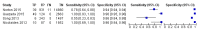






Update of
References
References to studies included in this review
Alberti 2015 {published and unpublished data}
-
- Alberti A, Salomon LJ, Lorc'h M, Couloux A, Bussieres L, Goupil S, et al. Non‐invasive prenatal testing for trisomy 21 based on analysis of cell‐free fetal DNA circulating in the maternal plasma. Prenatal Diagnosis 2015;35(5):471‐6. [PUBMED: 25643828] - PubMed
-
- NCT01118507. Trisomy of chromosome 21 diagnosis by high output sequencing. clinicaltrials.gov/show/NCT01118507 Date first received: 28 April 2010.
Ashoor 2012 {published data only}
-
- Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome‐selective sequencing of maternal plasma cell‐free DNA for first‐trimester detection of trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology 2012;206(4):322.e1‐5. [PUBMED: 22464073] - PubMed
Ashoor 2013 {published data only}
-
- Ashoor G, Syngelaki A, Wang E, Struble C, Oliphant A, Song K, et al. Trisomy 13 detection in the first trimester of pregnancy using a chromosome‐selective cell‐free DNA analysis method. Ultrasound in Obstetrics & Gynecology 2013;41(1):21‐5. [PUBMED: 22996646] - PubMed
Benachi 2015 {published data only}
-
- Benachi A, Letourneau A, Kleinfinger P, Senat MV, Gautier E, Favre R, et al. Cell‐free DNA analysis in maternal plasma in cases of fetal abnormalities detected on ultrasound examination. Obstetrics and Gynecology 2015;125(6):1330‐7. [PUBMED: 26000504] - PubMed
Bevilacqua 2015 {published data only}
-
- Bevilacqua E, Gil MM, Nicolaides KH, Ordonez E, Cirigliano V, Dierickx H, et al. Performance of screening for aneuploidies by cell‐free DNA analysis of maternal blood in twin pregnancies. Ultrasound in Obstetrics & Gynecology 2015;45(1):61‐6. [PUBMED: 25297464] - PubMed
Bianchi 2012 {published data only}
-
- Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP, et al. Genome‐wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstetrics and Gynecology 2012;119(5):890‐901. [PUBMED: 22362253] - PubMed
Bianchi 2013 {published and unpublished data}
-
- Bianchi DW, Prosen T, Platt LD, Goldberg JD, Abuhamad AZ, Rava RP, et al. Massively parallel sequencing of maternal plasma DNA in 113 cases of fetal nuchal cystichygroma. Obstetrics and Gynecology 2013;121(5):1057‐62. - PubMed
-
- NCT01122524. MatErnal bLood IS Source to accurately diagnose fetal Aneuploidy (MELISSA). clinicaltrials.gov/show/NCT01122524 Date first received: 11 May 2010.
Bianchi 2014a {published and unpublished data}
-
- Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, et al. DNA sequencing versus standard prenatal aneuploidy screening. New England Journal of Medicine 2014;370(9):799‐808. [PUBMED: 24571752] - PubMed
-
- NCT01663350. Comparison of aneuploidy risk evaluations. clinicaltrials.gov/show/NCT01663350 Date first received: 31 July 2012.
Bijok 2014 {published data only}
-
- Bijok J, Gorzelnik K, Massalska D, Ilnicka A, Pawlowska B, Zimowski JG, et al. Non‐invasive prenatal diagnosis of the most common aneuploidies with cell‐free fetal DNA in maternal serum‐‐preliminary results [Nieinwazyjna diagnostyka prenatalna najczestszych aneuploidii na podstawie plodowego DNA we krwi matki ‐‐doniesienie wstepne]. Ginekologia Polska 2014;85(3):208‐13. [PUBMED: 24783433] - PubMed
Canick 2012 {published data only}
-
- Canick JA, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Ehrich M, Boom D, et al. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenatal Diagnosis 2012;32(8):730‐4. [PUBMED: 22585317] - PubMed
Chen 2011 {published data only}
Chiu 2011 {published data only}
Comas 2015 {published data only}
-
- Comas C, Echevarria M, Rodriguez MA, Prats P, Rodriguez I, Serra B. Initial experience with non‐invasive prenatal testing of cell‐free DNA for major chromosomal anomalies in a clinical setting. Journal of Maternal‐fetal & Neonatal Medicine 2015;28(10):1196‐201. [PUBMED: 25048745] - PubMed
del Mar Gil 2014 {published data only}
-
- Mar Gil M, Quezada MS, Bregant B, Syngelaki A, Nicolaides KH. Cell‐free DNA analysis for trisomy risk assessment in first‐trimester twin pregnancies. Fetal Diagnosis and Therapy 2014;35(3):204‐11. [PUBMED: 24247435] - PubMed
Ehrich 2011 {published data only}
-
- Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. American Journal of Obstetrics and Gynecology 2011;204(3):205.e1‐11. [PUBMED: 21310373] - PubMed
Fiorentino 2016 {published data only}
-
- Fiorentino F, Bono S, Pizzuti F, Mariano M, Polverari A, Duca S, et al. The importance of determining the limit of detection of non‐invasive prenatal testing methods. Prenatal Diagnosis 2016;36(4):304‐11. [PUBMED: 26815144] - PubMed
Gil 2016 {published data only}
-
- Gil MM, Revello R, Poon LC, Akolekar R, Nicolaides KH. Clinical implementation of routine screening for fetal trisomies in the UK NHS: cell‐free DNA test contingent on results from first‐trimester combined test. Ultrasound in Obstetrics & Gynecology 2016;47(1):45‐52. [PUBMED: 26498918] - PubMed
Hall 2014 {published data only}
Hooks 2014 {published data only}
-
- Hooks J, Wolfberg AJ, Wang ET, Struble CA, Zahn J, Juneau K, et al. Non‐invasive risk assessment of fetal sex chromosome aneuploidy through directed analysis and incorporation of fetal fraction. Prenatal Diagnosis 2014;34(5):496‐9. [PUBMED: 24510887] - PubMed
Hou 2012 {published data only}
-
- Hou QF, Wu D, Chu Y, Kang B, Liao SX, Yang YL, et al. Clinical application of noninvasive prenatal diagnosis using cell free fetal DNA in maternal plasma. Zhonghua Fu Chan Ke za Zhi 2012;47(11):813‐7. [PUBMED: 23302120] - PubMed
Huang 2014 {published data only}
-
- Huang X, Zheng J, Chen M, Zhao Y, Zhang C, Liu L, et al. Noninvasive prenatal testing of trisomies 21 and 18 by massively parallel sequencing of maternal plasma DNA in twin pregnancies. Prenatal Diagnosis 2014;34(4):335‐40. [PUBMED: 24357023] - PubMed
Jackson 2014 {published data only}
-
- Jackson J, Hamar B, Lazar E, Lim K, Rodriguez D, Stock K, et al. Nuchal translucency measurement plus non‐invasive prenatal testing to screen for aneuploidy in a community‐based average‐risk population. Ultrasound in Obstetrics & Gynecology 2014; Vol. 44, issue 4:491. [PUBMED: 24890031] - PubMed
Jeon 2014 {published data only}
Jiang 2012 {published data only}
Johansen 2016 {published data only}
-
- Johansen P, Richter SR, Balslev‐Harder M, Miltoft CB, Tabor A, Duno M, et al. Open source non‐invasive prenatal testing platform and its performance in a public health laboratory. Prenatal Diagnosis 2016;36(6):530‐6. [PUBMED: 27027563] - PubMed
Ke 2015 {published data only}
Kim 2016 {published data only}
Korostelev 2014 {published data only}
-
- Korostelev S, Totchiev G, Kanivets I, Gnetetskaya V. Association of non‐invasive prenatal testing and chromosomal microarray analysis for prenatal diagnostics. Gynecological Endocrinology 2014;30:13‐6. - PubMed
Lau 2012 {published data only}
-
- Lau TK, Chen F, Pan X, Pooh RK, Jiang F, Li Y, et al. Noninvasive prenatal diagnosis of common fetal chromosomal aneuploidies by maternal plasma DNA sequencing. Journal of Maternal‐Fetal & Neonatal Medicine 2012;25(8):1370‐4. [PUBMED: 22070770] - PubMed
Lee 2015 {published data only}
Lefkowitz 2016 {published data only}
-
- Lefkowitz RB, Tynan JA, Liu T, Wu Y, Mazloom AR, Almasri E, et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. American Journal of Obstetrics and Gynecology 2016;215(2):227.e1‐227.e16. [PUBMED: 26899906] - PubMed
Liang 2013 {published data only}
-
- Liang D, Lv W, Wang H, Xu L, Liu J, Li H, et al. Non‐invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenatal Diagnosis 2013;33(5):409‐15. [PUBMED: 23299662] - PubMed
Liu 2012 {published data only}
-
- Liu HY, Wu D, Li H, Guo SK, Zhang CY, Liao SX, et al. Significance of detecting free DNA from maternal plasma for the diagnosis of fetal chromosomal aneuploidies. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi [Chinese journal of medical genetics] 2012;29(4):435‐8. [PUBMED: 22875501] - PubMed
Ma 2016 {published data only}
-
- Ma J, Wang Y, Wang W, Dong Y, Xu C, Zhou A, et al. Validation of combinatorial probe‐anchor ligation (cPAL) based sequencing method for non‐invasive prenatal testing in trisomy detection by a central laboratory. Ultrasound in Obstetrics & Gynecology 2016;50(1):49‐57. [PUBMED: 27363706] - PubMed
Mazloom 2013 {published data only}
-
- Mazloom AR, Dzakula Z, Oeth P, Wang H, Jensen T, Tynan J, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell‐free DNA from maternal plasma. Prenatal Diagnosis 2013;33(6):591‐7. [PUBMED: 23592550] - PubMed
Nicolaides 2012 {published data only}
-
- Nicolaides KH, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first‐trimester population. American Journal of Obstetrics and Gynecology 2012;207(5):374.e1‐6. [PUBMED: 23107079] - PubMed
Nicolaides 2013 {published data only}
-
- Nicolaides KH, Syngelaki A, Gil M, Atanasova V, Markova D. Validation of targeted sequencing of single‐nucleotide polymorphisms for non‐invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Prenatal Diagnosis 2013;33(6):575‐9. - PubMed
Nicolaides 2014a {published data only}
-
- Nicolaides KH, Musci TJ, Struble CA, Syngelaki A, Gil MM. Assessment of fetal sex chromosome aneuploidy using directed cell‐free DNA analysis. Fetal Diagnosis and Therapy 2014;35(1):1‐6. [PUBMED: 24335155] - PubMed
Norton 2012 {published data only}
-
- Norton ME, Brar H, Weiss J, Karimi A, Laurent LC, Caughey AB, et al. Non‐Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology 2012;207(2):137.e1‐8. [PUBMED: 22742782] - PubMed
Norton 2015 {published and unpublished data}
-
- NCT01511458. Non‐invasive chromosomal examination of Trisomy Study. clinicaltrials.gov/show/NCT01511458 Date first received: 13 January 2012.
-
- Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, et al. Cell‐free DNA analysis for noninvasive examination of trisomy. New England Journal of Medicine 2015;372(17):1589‐97. [PUBMED: 25830321] - PubMed
Palomaki 2012 {published data only}
-
- Palomaki GE, Deciu C, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genetics in Medicine 2012;14(3):296‐305. [PUBMED: 22281937] - PMC - PubMed
Papageorghiou 2016a {published data only}
Papageorghiou 2016b {published data only}
Pergament 2014 {published data only}
Persico 2016 {published data only}
-
- Persico N, Boito S, Ischia B, Cordisco A, Robertis V, Fabietti I, et al. Cell‐free DNA testing in the maternal blood in high‐risk pregnancies after first‐trimester combined screening. Prenatal Diagnosis 2016;36(3):232‐6. [PUBMED: 26749576] - PubMed
Poon 2016 {published data only}
Porreco 2014 {published and unpublished data}
-
- NCT00847990. Non‐Invasive screening for fetal aneuploidy. clinicaltrials.gov/show/NCT00847990 Date first received: 18 February 2009.
-
- Porreco RP, Garite TJ, Maurel K, Marusiak B, Ehrich M, Boom D, et al. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. American Journal of Obstetrics and Gynecology 2014;211(4):365.e1‐12. [PUBMED: 24657131] - PubMed
Quezada 2015 {published data only}
-
- Quezada MS, Gil MM, Francisco C, Orosz G, Nicolaides KH. Screening for trisomies 21, 18 and 13 by cell‐free DNA analysis of maternal blood at 10‐11 weeks' gestation and the combined test at 11‐13 weeks. Ultrasound in Obstetrics & Ggynecology 2015;45(1):36‐41. [PUBMED: 25251385] - PubMed
Samango‐Sprouse 2013 {published data only}
Sehnert 2011 {published data only}
-
- Sehnert AJ, Rhees B, Comstock D, Feo E, Heilek G, Burke J, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell‐free fetal DNA from maternal blood. Clinical Chemistry 2011;57(7):1042‐9. [PUBMED: 21519036] - PubMed
Shaw 2014 {published data only}
-
- Shaw SW, Hsiao CH, Chen CY, Ren Y, Tian F, Tsai C, et al. Noninvasive prenatal testing for whole fetal chromosomal aneuploidies: a multicenter prospective cohort trial in Taiwan. Fetal Diagnosis and Therapy 2014;35(1):13‐7. [PUBMED: 24296685] - PubMed
Song 2013 {published data only}
-
- Song Y, Liu C, Qi H, Zhang Y, Bian X, Liu J. Noninvasive prenatal testing of fetal aneuploidies by massively parallel sequencing in a prospective Chinese population. Prenatal Diagnosis 2013;33(7):700‐6. - PubMed
Song 2015 {published data only}
-
- Song Y, Huang S, Zhou X, Jiang Y, Qi Q, Bian X, et al. Non‐invasive prenatal testing for fetal aneuploidies in the first trimester of pregnancy. Ultrasound in Obstetrics & Gynecology 2015;45(1):55‐60. - PubMed
Sparks 2012a {published data only}
-
- Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell‐free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology 2012;206(4):319.e1‐9. [PUBMED: 22464072] - PubMed
Stumm 2014 {published data only}
-
- Stumm M, Entezami M, Haug K, Blank C, Wustemann M, Schulze B, et al. Diagnostic accuracy of random massively parallel sequencing for non‐invasive prenatal detection of common autosomal aneuploidies: a collaborative study in Europe. Prenatal Diagnosis 2014;34(2):185‐91. [PUBMED: 24222400] - PubMed
Sukhikh 2015 {published data only}
-
- Sukhikh GT, Karetnikova NA, Shubina ES, Baranova EE, Korostin DO, Ekimov AN, et al. Noninvasive prenatal diagnosis of aneuploidies by next‐generation sequencing (NGS) in a group of high‐risk women. Akusherstvo i Ginekologiia 2015;4:5‐10.
Sung‐Hee 2015 {published data only}
-
- Sung‐Hee H, Young‐Ho Y, Jae‐Song R, Myung‐Soo K, Young‐Jin K, Kyoung‐Ryul L. Noninvasive prenatal test for fetal chromosomal aneuploidies by massively parallel sequencing of cell‐free fetal DNA in maternal plasma: The first clinical experience in Korea. Journal of Genetic Medicine 2015;12(2):85‐91.
Tynan 2016 {published data only}
-
- Tynan JA, Kim SK, Mazloom AR, Zhao C, McLennan G, Tim R, et al. Application of risk score analysis to low‐coverage whole genome sequencing data for the noninvasive detection of trisomy 21, trisomy 18, and trisomy 13. Prenatal Diagnosis 2016;36(1):56‐62. [PUBMED: 26505614] - PubMed
Verweij 2013 {published data only}
-
- Verweij EJ, Jacobsson B, Scheltema PA, Boer MA, Hoffer MJ, Hollemon D, et al. European non‐invasive trisomy evaluation (EU‐NITE) study: a multicenter prospective cohort study for non‐invasive fetal trisomy 21 testing. Prenatal Diagnosis 2013;33(10):996‐1001. [PUBMED: 23794121] - PubMed
Wang 2014 {published data only}
-
- Wang S, Gao Z, Lu Y, Li Y, Jiang S, Wang L, et al. Detection of fetal chromosomal aneuploidy in pregnant women at advanced maternal age during the first trimester. Nan Fang Yi Ke da Xue Xue Bao [Journal of Southern Medical University] 2014;34(5):655‐8. [PUBMED: 24849430] - PubMed
Wang 2015a {published data only}
-
- Wang L, Meng Q, Tang X, Yin T, Zhang J, Yang S, et al. Maternal mosaicism of sex chromosome causes discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Taiwanese Journal of Obstetrics & Gynecology 2015;54(5):527‐31. [PUBMED: 26522104] - PubMed
Yao 2014 {published data only}
-
- Yao H, Jiang F, Hu H, Gao Y, Zhu Z, Zhang H, et al. Detection of fetal sex chromosome aneuploidy by massively parallel sequencing of maternal plasma DNA: initial experience in a Chinese hospital. Ultrasound in Obstetrics & Gynecology 2014;44(1):17‐24. [PUBMED: 24616044] - PubMed
Zhang 2016 {published data only}
Zhou 2014a {published data only}
-
- Zhou Q, Pan L, Chen S, Chen F, Hwang R, Yang X, et al. Clinical application of noninvasive prenatal testing for the detection of trisomies 21, 18, and 13: a hospital experience. Prenatal Diagnosis 2014;34(11):1061‐5. [PUBMED: 24899146] - PubMed
Zhou 2014b {published data only}
-
- Zhou Q, Pan L, Chen S, Chen F, Hwang R, Yang X, et al. Clinical application of noninvasive prenatal testing for the detection of trisomies 21, 18, and 13: a hospital experience. Prenatal Diagnosis 2014;34(11):1061‐5. [PUBMED: 24899146] - PubMed
References to studies excluded from this review
Anderson 2015 {published data only}
-
- Anderson B, Zhang K, Nguyen Q, Tsao D, Liu Y, Livingston K, et al. An automated, non‐invasive prenatal screening assay (NIPS) for trisomy 21,18,13 in singleton and twin gestations. International Journal of Gynecology and Obstetrics 2015;131:E264.
Anselem 2016 {published data only}
-
- Anselem O, Keroui S, Deput‐Rampon C, Chartier M, Costa JM, Goffinet F, et al. Analysis of cell‐free DNA in maternal blood for detection of fetal trisomy 21 in high‐risk population: Couples acceptance and grounds for refusal [Étude de l'ADN foetal dans le sang maternel pour la détection de la trisomie 21 en population à risque accru : adhésion des couples et motifs de refus]. Journal de Gynecologie, Obstetrique et Biologie de la Reproduction 2016;45(8):918‐23. [PUBMED: 26780844] - PubMed
Bayindir 2015 {published data only}
Beamon 2013 {published data only}
-
- Beamon C, Hardisty E, Harris S, Vora N. Promises and pitfalls of a new technology: a single center experience with noninvasive prenatal testing (NIPT). American Journal of Obstetrics and Gynecology 2013;208(1):S244‐S5.
Beamon 2014 {published data only}
-
- Beamon CJ, Hardisty EE, Harris SC, Vora NL. A single center's experience with noninvasive prenatal testing. Genetics in Medicine 2014;16(9):681‐7. [PUBMED: 24675675] - PubMed
Belloin 2016 {published data only}
-
- Belloin C, Jacquemard F, Bernabe‐Dupont C, Viot G, Lohmann L, Grange G. The noninvasive prenatal testing for Down's Syndrome. Retrospective study of 8821 patients [Le dépistage prénatal non invasif de la trisomie 21. Étude rétrospective à propos de 8821 patientes]. Journal de Gynecologie, Obstetrique et Biologie de la Reproduction 2016;45(9):1127‐32. [PUBMED: 27091545] - PubMed
Benachi 2015b {published data only}
-
- Benachi A, Costa JM. Non invasive prenatal diagnosis of trisomy 21. La Revue du Praticien 2015;65(2):160‐2. [PUBMED: 25939212] - PubMed
Benachi 2016 {published data only}
-
- Benachi A, Letourneau A, Kleinfinger P, Senat MV, Gautier E, Favre R, et al. Performance and indications of noninvasive prenatal testing using cell free circulating fetal DNA (cffDNA) for the detection of fetal trisomy 21, 18 and 13 in France [Performance et indication du dépistage des trisomies 21, 18 et 13 en France par l'analyse de l'ADN foetal dans le sang maternel.]. Journal de Gynécologie, Obstétrique et Biologie de la Reproduction 2016;45(6):633‐40. [PUBMED: 26518155] - PubMed
Benn 2015 {published data only}
-
- Benn P. Re: Non‐invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146 958 pregnancies. Zhang H, Gao Y, Jiang F, Fu M, Yuan Y, Guo Y, et al. Ultrasound in Obstetrics & Gynecology 2015; 45(5): 530‐8.. Ultrasound in Obstetrics & Gynecology 2015;45(5):512‐3. [PUBMED: 25914392] - PubMed
Bhatt 2014 {published data only}
-
- Bhatt S, Parsa S, Snyder H, Taneja P, Halks‐Miller M, Seltzer W, et al. Clinical laboratory experience with noninvasive prenatal testing: Update on clinically relevant metrics. Prenatal Diagnosis 2014;34(Suppl 1):48.
Bianchi 2012a {published data only}
-
- Bianchi D, Platt L, Goldberg J, Abuhamad A, Sehnert A, Rava R. Whole genome maternal plasma DNA sequencing accurately detects autosomal and sex chromosome aneuploidies. Prenatal Diagnosis 2012;32((Bianchi D.) Tufts Medical Center, Mother Infant Research Institute, Boston, United States):3‐4.
Bianchi 2014b {published data only}
-
- Bianchi DW, Lamar Parker R, Wentworth J, Madankumar R, Saffer C, Das AF, et al. DNA sequencing versus standard prenatal aneuploidy screening. Obstetrical and Gynecological Survey 2014;69(6):319‐21. [EMBASE: 2014389701]
Bianchi 2014c {published data only}
-
- Bianchi D, Swanson A, Parsa S, Bhatt S, Halks‐Miller M, Sehnert A, et al. NIPT for sex chromosome aneuploidy: Initial clinical laboratory experience and biologic reasons for discordant results. American Journal of Obstetrics and Gynecology 2014;210(1):S87‐S8.
Bianchi 2015a {published data only}
-
- Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 2015;314(2):162‐9. [PUBMED: 26168314] - PubMed
Bianchi 2015b {published data only}
-
- Bianchi DW, Parsa S, Bhatt S, Halks‐Miller M, Kurtzman K, Sehnert AJ, et al. Fetal sex chromosome testing by maternal plasma DNA sequencing: clinical laboratory experience and biology. Obstetrics and Gynecology 2015;125(2):375‐82. - PubMed
Bianchi 2015c {published data only}
-
- Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, et al. Noninvasive prenatal yesting and incidental detection of occult maternal malignancies. Obstetrical and Gynecological Survey 2015;70(12):744‐6. - PubMed
Bianchi 2015d {published data only}
-
- Bianchi D, Chudova D, Sehnert A, Bhatt S, Murray K, Prosen T, et al. Incidental detection of occult maternal malignancies by noninvasive prenatal testing. Prenatal Diagnosis 2015;35:10‐1. - PubMed
BlueCross BlueShield Asssociation 2014 {published data only}
-
- BlueCross BlueShield Asssociation. Noninvasive prenatal cell‐free fetal DNA‐based screening for aneuploidies other than trisomy 21. Technology Evaluation Center Assessment Program Executive summary 2014;29(7):1‐7. [PUBMED: 25577816] - PubMed
Brady 2016 {published data only}
-
- Brady P, Brison N, Bogaert K, Ravel T, Peeters H, Esch H, et al. Clinical implementation of NIPT ‐ technical and biological challenges. Clinical Genetics 2016;89(5):523‐30. [PUBMED: 25867715] - PubMed
Chen 2013 {published data only}
-
- Chen F, Zheng J, Chen M, Fang Q, Wei Y, Zhang C, et al. Noninvasive prenatal testing of Trisomy 21 and 18 by maternal plasma sequencing in twin pregnancies. Prenatal Diagnosis 2013;33:14.
Chen 2014 {published data only}
-
- Chen M, Ma GC, Yeang CH. Genome‐wide normalized score: A novel counting algorithm to detect fetal trisomies in non‐invasive prenatal testing. Prenatal Diagnosis 2014;34:49. - PubMed
Cherry 2014 {published data only}
-
- Cherry AM, Williams JM Jr, Manning MA. Accuracy of non‐invasive prenatal screening for trisomies 13, 18 and 21 and sex chromosome aneuploidy. Cytogenetic and Genome Research 2014;142(3):240‐1.
Cheung 2015 {published data only}
-
- Cheung SW, Patel A, Leung TY. Accurate description of DNA‐based noninvasive prenatal screening. New England Journal of Medicine 2015; Vol. 372, issue 17:1675‐7. [PUBMED: 25830325] - PubMed
Chiu 2008 {published data only}
-
- Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proceedings of the National Academy of Sciences of the United States of America 2008;105(51):20458‐63. [PUBMED: 19073917] - PMC - PubMed
Chiu 2010 {published data only}
-
- Chiu RW, Sun H, Akolekar R, Clouser C, Lee C, McKernan K, et al. Maternal plasma DNA analysis with massively parallel sequencing by ligation for noninvasive prenatal diagnosis of trisomy 21. Clinical Chemistry 2010;56(3):459‐63. [PUBMED: 20026875] - PubMed
Christina 2012 {published data only}
Cinnioglu 2012 {published data only}
-
- Cinnioglu C. The application of bioinformatics to genetic testing for the detection of human aneuploidy and genotyping. Reproductive BioMedicine Online 2012;24:S37.
Cirigliano 2013 {published data only}
-
- Cirigliano V, Ordonez E, Rueda L, Moreno M, Palao B, Paz Canadas M. Introduction of cfDNA based screening for common trisomies in Spain. Prenatal Diagnosis 2013;33:85‐6.
Cirigliano 2014 {published data only}
-
- Cirigliano V, Ordoñez E, Rueda L, Cañadas P, Moreno M, Palao B. cfDNA based aneuploidy screening in Spain: Results of one year clinical application. Prenatal Diagnosis 2014;34:51.
Cuckle 2015 {published data only}
-
- Cuckle H, Benn P, Pergament E. Cell‐free DNA screening for fetal aneuploidy as a clinical service. Clinical Biochemistry 2015;48(15):932‐41. [PUBMED: 25732593] - PubMed
Curnow 2014 {published data only}
-
- Curnow K, Ryan A, Banjevic M, Shchegrova S, Babiarz J, Constantin T, et al. Observations following clinical implementation of a cfDNA and single‐nucleotide polymorphism‐based non‐invasive prenatal aneuploidy test (NIPT). Prenatal Diagnosis 2014;34:51‐2.
Dan 2012 {published data only}
-
- Dan S, Wang W, Ren J, Li Y, Hu H, Xu Z, et al. Clinical application of massively parallel sequencing‐based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenatal Diagnosis 2012;32(13):1225‐32. [PUBMED: 23138752] - PubMed
Dar 2014 {published data only}
-
- Dar P, Curnow KJ, Gross SJ, Hall MP, Stosic M, Demko Z, et al. Clinical experience and follow‐up with large scale single‐nucleotide polymorphism‐based noninvasive prenatal aneuploidy testing. American Journal of Obstetrics and Gynecology 2014;211(5):527.e1‐527.e17. [PUBMED: 25111587] - PubMed
De Ligt 2013 {published data only}
-
- Ligt J, Janssen I, Bon B, Buysse K, Gomes I, Eggink A, et al. Detecting partial fetal aneuploidies by MPS: An unexpected discrepancy between amniotic fluid and ccffDNA. Prenatal Diagnosis 2013;33(Suppl 1):72‐3. [ISSN 01973851]
Denona 2016 {published data only}
-
- Denona B, Mone F, Cathcart B, Mahony R, Carroll S, Higgins S, et al. Changing rates of noninvasive prenatal testing and prenatal invasive testing in a tertiary centre. BJOG: an international journal of obstetrics and gynaecology 2016;123:75‐6.
Discenza 2015 {published data only}
-
- Discenza M, Ahern D, Coles K, Dobson L, Reiss R. Sex chromosome aneuploidy detection by NIPT: helpful or hazardous?. Prenatal Diagnosis 2015;35:105‐6. - PubMed
Dobson 2015 {published data only}
-
- Dobson L, Reiff E, Little S, Bromley B, Wilkins‐Haug L. Clinical course and patient outcomes following positive noninvasive prenatal testing (NIPT). Prenatal Diagnosis 2015;35:3. - PubMed
Dobson 2016 {published data only}
-
- Dobson LJ, Reiff ES, Little SE, Wilkins‐Haug L, Bromley B. Patient choice and clinical outcomes following positive noninvasive prenatal screening for aneuploidy with cell‐free DNA (cfDNA). Prenatal Diagnosis 2016;36(5):456‐62. [PUBMED: 26938930] - PubMed
Dong 2016 {published data only}
-
- Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian Q, et al. Low‐pass whole‐genome sequencing in clinical cytogenetics: a validated approach. Genetics in Medicine 2016;18(9):940‐8. [PUBMED: 26820068] - PubMed
Duenwald 2016 {published data only}
-
- Duenwald S, Barbacioru C, Deciu C, Chen G, Comstock D, Skvortsov D, et al. Development of a novel paired‐end sequencing‐based noninvasive prenatal test. American Journal of Obstetrics and Gynecology 2016;214(1):S254‐5.
Ehrich 2011a {published data only}
-
- Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Obstetrical & Gynecological Survey 2011;66(6):342‐4. - PubMed
Eiben 2014 {published data only}
-
- Eiben B, Glaubitz R, Kagan KO. Non‐invasive prenatal diagnostics: ETS and NGS‐based tests. Medizinische Genetik 2014;26(4):382‐90. [EMBASE: 2014948590]
Ellison 2015 {published data only}
-
- Ellison C, Sun Y, Hogg G, Fox J, Tao H, McCarthy E, et al. Leveraging targeted sequencing of paired homologs for noninvasive detection of fetal aneuploidies. Prenatal Diagnosis 2015;35:54.
Faas 2011 {published data only}
-
- Faas B, Vissers L, Janssen I, Ligt J, Eggink A, Veltman J, et al. Multiplex massively parallel sequencing for noninvasive prenatal diagnosis. Chromosome Research 2011;19:S35‐S6.
Faas 2012 {published data only}
-
- Faas BH, Ligt J, Janssen I, Eggink AJ, Wijnberger LD, Vugt JM, et al. Non‐invasive prenatal diagnosis of fetal aneuploidies using massively parallel sequencing‐by‐ligation and evidence that cell‐free fetal DNA in the maternal plasma originates from cytotrophoblastic cells. Expert Opinion on Biological Therapy 2012;12 Suppl 1:S19‐26. [PUBMED: 22500971] - PubMed
Fairbrother 2013a {published data only}
-
- Fairbrother G, Johnson S, Musci TJ, Song K. Clinical experience of noninvasive prenatal testing with cell‐free DNA for fetal trisomies 21, 18, and 13, in a general screening population. Prenatal Diagnosis 2013;33(6):580‐3. [PUBMED: 23494956] - PubMed
Fairbrother 2013b {published data only}
-
- Fairbrother G, Johnson S, Musci T, Song K. Clinical experience of Harmony™ Prenatal Test, a noninvasive prenatal test, in a general screening population. Prenatal Diagnosis 2013;33:78. - PubMed
Fan 2008 {published data only}
Fang 2015 {published data only}
-
- Fang Y, Wang G, Wang C, Suo F, Gu M, Xia Y. The diagnosis pattern of mid‐trimester fetal chromosomal aneuploidy in Xuzhou and the clinical applications. Cell Biochemistry and Biophysics 2015;73(2):267‐70. [PUBMED: 25733332] - PubMed
Ferres 2013 {published data only}
-
- Ferres MA, Lichten L, Sachs A, Lau KM, Bianchi DW. Early experience with noninvasive DNA testing for aneuploidy in prenatal care. Prenatal Diagnosis 2013;33:73.
Fiorentino 2015 {published data only}
-
- Fiorentino F, Spinella F, Bono S, Pizzuti F, Mariano M, Polverari A, et al. Feasibility of noninvasive prenatal testing for common fetal aneuploidies in maternal serum with low levels circulating fetal cell‐free DNA fraction. Prenatal Diagnosis 2015;35:1.
Fosler 2015 {published data only}
-
- Fosler L, Winters P, Jones K, Platt L. Clinical laboratory experience with noninvasive prenatal testing in twin gestations. American Journal of Obstetrics and Gynecology 2015;212(1):S330‐S1.
Futch 2013 {published data only}
Gabriel 2014 {published data only}
-
- Gabriel, H.Biskup, S. Parallel prenatal testing for trisomies (‐13,‐18 and‐21), turner syndrome, 22q11.2 del/dup‐syndromes (e.g. DiGeorge), Smith‐Lemli‐Opitz syndrome and Noonan syndrome in fetuses with increased nuchal translucency using the AmpliSeq™ technology (Life Tech). Medizinische Genetik 2014;26(1):192‐3. [ISSN 09365931]
Galea 2014 {published data only}
-
- Galea M, Grehan S, Ives D, Evans K, Harraway J. A one‐year experience of non‐invasive prenatal testing in an Australian private pathology setting. Prenatal Diagnosis 2014;34:54.
Gao 2014 {published data only}
-
- Gao Y, Xie B, Liu R. Delivering noninvasive prenatal testing in a clinical setting using semiconductor sequencing platform. Science China Life Sciences 2014;57(7):737‐8. [PUBMED: 24969704] - PubMed
Gao 2015 {published data only}
-
- Gao Y, Zeng F, Fu M, Zhang H, Jiang F, Chen F, et al. Comparison of NIPT clinical performance in 72,382 high risk pregnant women and 40,287 low‐risk pregnant women. Prenatal Diagnosis 2015;35:2.
Geifman‐Holtzman 2013 {published data only}
-
- Geifman‐Holtzman O, Berman J, Ness AL, Cohen A, Fischer R, Carre A, et al. NIPDT registry‐a critical utilization of non‐invasive prenatal diagnosis test recently introduced to clinical practice. American Journal of Obstetrics and Gynecology 2013;208(1):S254.
Geifman‐Holtzman 2014 {published data only}
-
- Geifman‐Holtzman O, Berman J, Xiong Y, Carre A, Ness A, Fang YMV, et al. Noninvasive prenatal testing (NIPT) registry patients' results and providers' perspective. American Journal of Obstetrics and Gynecology 2014;210(1):S96.
Gerundino 2017 {published data only}
-
- Gerundino F, Giachini C, Contini E, Benelli M, Marseglia G, Giuliani C, et al. Validation of a method for noninvasive prenatal testing for fetal aneuploidies risk and considerations for its introduction in the Public Health System. Journal of Maternal‐Fetal & Neonatal Medicine 2017;30(6):710‐6. [PUBMED: 27226231] - PubMed
Gil 2013 {published data only}
-
- Gil MM, Quezada MS, Bregant B, Ferraro M, Nicolaides KH. Implementation of maternal blood cell‐free DNA testing in early screening for aneuploidies. Ultrasound in Obstetrics & Gynecology 2013;42(1):34‐40. [PUBMED: 23744609] - PubMed
Gil 2015 {published data only}
-
- Gil MM, Giunta G, Macalli EA, Poon LC, Nicolaides KH. UK NHS pilot study on cell‐free DNA testing in screening for fetal trisomies: factors affecting uptake. Ultrasound in Obstetrics & Gynecology 2015;45(1):67‐73. [PUBMED: 25302655] - PubMed
Gnetetskaya 2015 {published data only}
-
- Gnetetskaya V, Kurtser M. Diagnosis of chromosomal aneuploidies using non‐invasive prenatal test in Moscow. Journal of Perinatal Medicine 2015;43(s1):1300‐64.
Grati 2014 {published data only}
-
- Grati FR, Malvestiti F, Ferreira JC, Bajaj K, Gaetani E, Agrati C, et al. Fetoplacental mosaicism: potential implications for false‐positive and false‐negative noninvasive prenatal screening results. Genetics in Medicine 2014;16(8):620‐4. [PUBMED: 24525917] - PubMed
Gray 2013 {published data only}
-
- Gray KJ, Wilkins‐Haug L, Gerrol P, Reiss R. Introduction of ffDNA testing for aneuploidy screening: Indications, changes in invasive testing rates, and caveats. Prenatal Diagnosis 2013;33:74.
Gromminger 2014 {published data only}
Guex 2013 {published data only}
-
- Guex N, Iseli C, Syngelaki A, Deluen C, Pescia G, Nicolaides KH, et al. A robust second‐generation genome‐wide test for fetal aneuploidy based on shotgun sequencing cell‐free DNA in maternal blood. Prenatal Diagnosis 2013;33(7):707‐10. - PubMed
Halks‐Miller 2015 {published data only}
-
- Halks‐Miller M, Chudova D, Bianchi DW. Maternal malignancies detected with noninvasive prenatal testing‐‐reply. JAMA 2015; Vol. 314, issue 20:2192‐3. [PUBMED: 26599192] - PubMed
Harasim 2016 {published data only}
-
- Harasim T, Wagner A, Rath S, Behaqi P, Rost I, Klein H‐G. Prenatalis® NIPT: Accredited high resolution non‐invasive prenatal testing by using massive parallel ultra‐deep sequencing. Medizinische Genetik 2016;28(1):152‐3.
Hernandez‐Gomez 2015 {published data only}
-
- Hernandez‐Gomez M, Ramirez‐Arroyo E, Melendez‐Hernandez R, Garduno‐Zaraza LM, Mayen‐Molina DG. Non invasive prenatal test (NIPT) in maternal blood by parallel massive sequencing. Initial experience in Mexican women and literature review [Prueba prenatal no invasiva (NIPT) en sangre materna a traves de secuenciacion masiva paralela (MPS). Experiencia inicial en mujeres mexicanas y revision de la bibliografia]. Ginecologia y Obstetricia de Mexico 2015;83(5):277‐88. [PUBMED: 26233973] - PubMed
Hofmann 2013 {published data only}
-
- Hofmann W, Entezami M, Haug K, Blank C, Wustemann M, Schulze B, et al. Diagnostic accuracy for the noninvasive prenatal detection of common autosomal aneuploidies. Prenatal Diagnosis 2013;33:75. - PubMed
Hofmann 2014 {published data only}
-
- Hofmann W, Grömminger S, Schöck U, Bonnet J, Smerdka P. Non‐invasive prenatal testing (NIPT): Laboratory experiences of PrenaTest®. Medizinische Genetik 2014;26(1):194.
Hofmann 2015 {published data only}
-
- Hofmann W, Grömminger S, Sachse M, Bonnet J, Schöck U. Recent bioinformatic advances of non‐invasive prenatal detection to enhance diagnostic accuracy and aneuploidy discovery. Prenatal Diagnosis 2015;35:57.
Hu 2014 {published data only}
-
- Hu Y, Duan H, Li J, Zhu R. Study of clinical indications for non‐invasive prenatal testing based on distributions of fetal chromosomal abnormalities. Prenatal Diagnosis 2014;34:59.
Hu 2015 {published data only}
-
- Hu HJ, Kwon YJ, Oh M, Kim J, Cho DY, Seo DH. Evaluating the results of the Momguard™ noninvasive prenatal test. Journal of Genetic Medicine 2015;12(2):96‐9. [DOI: 10.5734/JGM.2015.12.2.96] - DOI
Hui 2015a {published data only}
-
- Hui L, Teoh M, Silva Costa F, Ramsay P, Palma‐Dias R, Richmond Z, et al. Clinical implementation of noninvasive prenatal testing by Australian sonologists. BJOG: a international journal of obstetrics and gynaecology 2015;122:52.
Hui 2015b {published and unpublished data}
-
- Hui L, Teoh M, Silva C, Ramsay P, Palma‐Dias R, Richmond Z, et al. Clinical implementation of cell‐free DNA‐based aneuploidy screening: perspectives from a national audit. Ultrasound in Obstetrics & Gynecology 2015;45(1):10‐5. - PubMed
Jackson 2013 {published data only}
-
- Jackson J, Dever K, Hamar B, Lazar E, Lim KH, Rodriguez D, et al. Nuchal translucency plus noninvasive prenatal testing screening for aneuploidy in a population of low‐and high‐risk patients. Prenatal Diagnosis 2013;33:22.
Jensen 2013 {published data only}
Jensen 2015 {published data only}
-
- Jensen TJ, Deciu C, Ehrich M, Geis J, Kim SK, Tao H. Selective enrichment of genomic loci for the noninvasive detection of fetal aneuploidies. Obstetrics and Gynecology 2015;125:92S.
Jin 2014 {published data only}
-
- Jin Y, Miao Z, Ge J, Zhang W, Li S, Liu X. Prenatal diagnosis of fetal chromosome aneuploidy by massively parallel genomic sequencing. National Medical Journal of China 2014;94(23):1788‐90. - PubMed
Johnson 2013 {published data only}
-
- Johnson J, Pastuck M, Metcalfe A, Connors G, Krause R, Wilson D, et al. First‐trimester Down syndrome screening using additional serum markers with and without nuchal translucency and cell‐free DNA. Prenatal Diagnosis 2013;33(11):1044‐9. - PubMed
Juneau 2014 {published data only}
-
- Juneau K, Bogard PE, Huang S, Mohseni M, Wang ET, Ryvkin P, et al. Microarray‐based cell‐free DNA analysis improves noninvasive prenatal testing. Fetal Diagnosis and Therapy 2014;36(4):282‐6. - PubMed
Kagan 2015 {published data only}
Kalantar 2014 {published data only}
-
- Kalantar SM, Sitar G, Carabresy J. Is time for using non‐invasive pre‐natal diagnosis for genetic disorders?. Iranian Journal of Reproductive Medicine 2014;12(6):15.
Karlsson 2015 {published data only}
-
- Karlsson K, Sahlin E, Iwarsson E, Westgren M, Nordenskjold M, Linnarsson S. Amplification‐free sequencing of cell‐free DNA for prenatal non‐invasive diagnosis of chromosomal aberrations. Genomics 2015;105(3):150‐8. - PubMed
Kershberg 2015 {published data only}
-
- Kershberg H, Greenberg J, Liou A, Logg A, Alvarado M, Natoli J, et al. Positive and negative predictive values of NIPT in an integrated health care system: summary of NIPT outcomes in Kaiser Permanente Southern California. Prenatal Diagnosis 2015;35:58.
Kinde 2012 {published data only}
Korabecna 2012 {published data only}
-
- Korabecna M. Non invasive prenatal diagnostics of most frequent chromosomal aneuploidies in the language of numbers. Aktualni Gynekologie a Porodnictvi 2012;4(1):114‐5.
Koumbaris 2016 {published data only}
-
- Koumbaris G, Kypri E, Tsangaras K, Achilleos A, Mina P, Neofytou M, et al. Cell‐free DNA analysis of targeted genomic regions in maternal plasma for non‐invasive prenatal testing of trisomy 21, trisomy 18, trisomy 13, and fetal sex. Clinical Chemistry 2016;62(6):848‐55. [PUBMED: 27117469] - PubMed
Kurtser 2015 {published data only}
-
- Kurtser MA, Gnetetskaya VA. A noninvasive prenatal test in the diagnosis of chromosome aneuploidies. Akushersivo i Ginekologiya 2015;8:65‐9.
Lambert‐Messerlian 2014 {published data only}
-
- Lambert‐Messerlian G, Kloza EM, Williams J 3rd, Loucky J, O'Brien B, Wilkins‐Haug L, et al. Maternal plasma DNA testing for aneuploidy in pregnancies achieved by assisted reproductive technologies. Genetics in Medicine 2014;16(5):419‐22. [PUBMED: 24091801] - PubMed
Larion 2015 {published data only}
-
- Larion S, Warsof S, Romary L, Mlynarczyk M, Abuhamad A. Three year clinical experience with noninvasive prenatal testing in 3000 high risk cases in the United States. Prenatal Diagnosis 2015;35:59.
Lau 2012a {published data only}
Lau 2013 {published data only}
-
- Lau TK, Jiang F, Chan MK, Zhang H, Lo PS, Wang W. Non‐invasive prenatal screening of fetal Down syndrome by maternal plasma DNA sequencing in twin pregnancies. Journal of Maternal‐Fetal & Neonatal Medicine 2013;26(4):434‐7. [PUBMED: 23035860] - PubMed
Lau 2014 {published data only}
-
- Lau TK, Cheung SW, Lo PS, Pursley AN, Chan MK, Jiang F, et al. Non‐invasive prenatal testing for fetal chromosomal abnormalities by low‐coverage whole‐genome sequencing of maternal plasma DNA: review of 1982 consecutive cases in a single center. Ultrasound in Obstetrics & Gynecology 2014;43(3):254‐64. [PUBMED: 24339153] - PubMed
Lebo 2015 {published data only}
Leung 2013 {published data only}
-
- Leung TY, Qu JZZ, Liao GJW, Jiang P, Cheng YKY, Chan KCA, et al. Noninvasive twin zygosity assessment and aneuploidy detection by maternal plasma DNA sequencing. Prenatal Diagnosis 2013;33(7):675‐81. - PubMed
Levandoski 2015 {published data only}
-
- Levandoski K, Marchand K, Briere L, Ferres M. One clinic's experience with discordant NIPT results for trisomies 18 and 13: Practical and psychosocial implications of non‐invasive prenatal testing. Prenatal Diagnosis 2015;35:106‐7.
Levy 2013 {published data only}
-
- Levy B, Banjevic M, Hill M, Zimmermann B, Ryan A, Sigurjonsson S, et al. Targeted sequencing of SNPs results in highly accurate noninvasive detection of fetal aneuploidy of chromosomes 13, 18, 21, X, and Y: A validation study. Human Reproduction 2013;28:i26‐i7.
Levy 2013a {published data only}
-
- Levy B, Banjevic M, Hill M, Zimmermann B, Ryan A, Sigurjonsson S, et al. Targeted sequencing of single‐nucleotide polymorphisms for accurate, noninvasive fetal aneuploidy detection at autosomal and sex chromosomes. Prenatal Diagnosis 2013;33:76.
Levy 2013b {published data only}
-
- Levy, Ryan B, Banjevic A, Hill M, Zimmermann M, Sigurjonsson B, et al. Highly multiplexed single‐nucleotide polymorphism amplification and sequencing to identify fetal aneuploidy from maternal cell‐free DNA. Journal of Perinatal Medicine 2013;41(Suppl 1):34‐218. [ISSN 03005577]
Li 2012 {published data only}
-
- Li X, Sehnert A, Rava R. Percentage of fetal cell‐free DNA in maternal plasma: Dependence on multiple clinical factors. Prenatal Diagnosis 2012;32:6.
Li 2015 {published data only}
-
- Li WH, Wang PH, Chuang CM, Chang YW, Yang MJ, Chen CY, et al. Noninvasive prenatal testing for fetal trisomy in a mixed risk factors pregnancy population. Taiwanese Journal of Obstetrics & Gynecology 2015;54(2):122‐5. [PUBMED: 25951714] - PubMed
Liao 2011 {published data only}
-
- Liao GJ, Lun FM, Zheng YW, Chan KC, Leung TY, Lau TK, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clinical Chemistry 2011;57(1):92‐101. [PUBMED: 21078840] - PubMed
Liao 2012 {published data only}
Liao 2013 {published data only}
-
- Liao C, Fu YG, Huang SY, Fu F, Xie GE. Rapid noninvasive prenatal diagnosis of Down syndrome with Ion Proton™. Prenatal Diagnosis 2013;33:76‐7.
Liao 2014 {published data only}
Liao 2014a {published data only}
-
- Liao C, Zhengfeng X, Zhang K. DNA sequencing versus standard prenatal aneuploidy screening. New England Journal of Medicine 2014; Vol. 371, issue 6:577‐8. [PUBMED: 25099588] - PubMed
Liu 2015 {published data only}
-
- Liu J, Wang H, Xi H, Jia Z, Zhou Y, Wu L. Application of next‐generation DNA sequencing for prenatal testing of fetal chromosomal aneuploidies. Zhonghua Yi Xue Yi Chuan Xue za Zhi [Chinese Journal of Medical Genetics] 2015;32(4):533‐7. [PUBMED: 26252102] - PubMed
Lo 2014 {published data only}
Lo 2014a {published data only}
-
- Lo K, Boustred C, McKay F, Fielding S, Plagnol V, Chitty L. A comparison of statistical methods for NIPT for aneuploidies and development of the RAPID‐R package. Prenatal Diagnosis 2014;34:59‐60.
Loucký 2013 {published data only}
-
- Loucký J, Zemánek M. Non invasive prenatal testing of most frequent chromosomal aneuploidies ‐ Some other aspects [Neinvazivní prenatální testóvání nejčastějsích chromozomálních aneuploidií ‐ Některé dalši aspekty]. Actual Gynecology and Obstetrics 2013;5(1):6‐7.
Louis‐Jacques 2014 {published data only}
-
- Louis‐Jacques A, Burans C, Robinson S, Schofield E, Smulian J, Rochon M. Use of commercial tests for aneuploidy screening using cell‐free fetal DNA in clinical practice. Obstetrics and Gynecology 2014;123(Suppl 1):154S.
Ma 2015 {published data only}
-
- Ma J, Pan H, Fu J, Yu L, Yang H. Perspective study of non‐invasive prenatal testing using cell‐free fetal DNA in high‐risk population. Zhonghua Yi Xue za Zhi 2015;95(11):849‐52. [PUBMED: 26080919] - PubMed
Ma 2015a {published data only}
-
- Ma J, Yang H, Chen F, Wang Y. Nationwide evaluation on noninvasive prenatal testing through maternal plasma DNA sequencing under governmental regulation in China. Prenatal Diagnosis 2015;35:67.
Manotaya 2016 {published data only}
-
- Manotaya S, Xu H, Uerpairojkit B, Chen F, Charoenvidhya D, Liu H, et al. Clinical experience from Thailand: noninvasive prenatal testing as screening tests for trisomies 21, 18 and 13 in 4736 pregnancies. Prenatal Diagnosis 2016;36(3):224‐31. [PUBMED: 26748603] - PubMed
Marchili 2015 {published data only}
-
- Marchili JP, Chaulet A, Guetmonovitch O, Margulies ND, Solarz V. One clinic's experience with cell free fetal DNA testing in Argentina. Prenatal Diagnosis 2015;35:60.
Mayen 2015 {published data only}
-
- Mayen DG, Hernandez‐Gomez M, Ramirez E, Meléndez R, Garduño LM. Noninvasive prenatal test, one year of experience in the genetics clinic at the Hospital Angeles Lomas in Mexico City. Prenatal Diagnosis 2015;35:60‐1.
Mazloom 2013a {published data only}
-
- Mazloom A, Dzakula Z, Wang H, Oeth P, Jensen T, Tynan J, et al. Detection of fetal sex chromosomal aneuploidies by sequencing circulating cell‐free DNA from maternal plasma. Prenatal Diagnosis 2013;33:77. - PubMed
McCullough 2014 {published data only}
McCullough 2014a {published data only}
-
- McCullough R, Almasri E, Guan X, Oeth P, Bombard A, Saldivar JS. Noninvasive prenatal testing: 100,000 patients clinical impact. American Journal of Obstetrics and Gynecology 2014;210(1):S58.
McCullough 2015 {published data only}
-
- McCullough R, Almasri E, Boomer T, Wardrop J, Oeth P, Paxton W, et al. High volume clinical laboratory noninvasive prenatal testing: A synopsis (or summary) of results from >375,000 patients. Prenatal Diagnosis 2015;35:51‐2.
McLennan 2016 {published data only}
-
- McLennan A, Palma‐Dias R, Silva Costa F, Meagher S, Nisbet DL, Scott F. Noninvasive prenatal testing in routine clinical practice‐‐an audit of NIPT and combined first‐trimester screening in an unselected Australian population. Australian & New Zealand Journal of Obstetrics & Gynaecology 2016;56(1):22‐8. [PUBMED: 26817523] - PubMed
Meck 2014 {published data only}
-
- Meck JM, Kramer DE, Aviram A, Trunca C, Riethmaier D, Pineda AD, et al. Non‐invasive prenatal screening from viewpoint of the cytogenetics laboratory. Cytogenetic and Genome Research 2014;142(3):241.
Meck 2015 {published data only}
-
- Meck JM, Kramer Dugan E, Matyakhina L, Aviram A, Trunca C, Pineda‐Alvarez D, et al. Noninvasive prenatal screening for aneuploidy: positive predictive values based on cytogenetic findings. American Journal of Obstetrics and Gynecology 2015;213(2):214.e1‐5. [PUBMED: 25843063] - PubMed
Meck 2015a {published data only}
-
- Meck J, Dugan EK, Pineda‐Alvarez D, Wray A, Richard G, Matyakhina L. Comparison of results from noninvasive prenatal screening and diagnostic cytogenetic testing. Prenatal Diagnosis 2015;35:61.
Mennuti 2015 {published data only}
-
- Mennuti MT, Chandrasekaran S, Khalek N, Dugoff L. Cell‐free DNA screening and sex chromosome aneuploidies. Prenatal Diagnosis 2015;35(10):980‐5. [PUBMED: 26088741] - PubMed
Minarik 2015 {published data only}
-
- Minarik G, Repiska G, Hyblova M, Nagyova E, Soltys K, Budis J, et al. Utilization of Benchtop Next Generation Sequencing Platforms Ion Torrent PGM and MiSeq in noninvasive prenatal testing for chromosome 21 trisomy and testing of impact of in silico and physical size selection on its analytical performance. PLOS One 2015;10(12):e0144811. [PUBMED: 26669558] - PMC - PubMed
Miron 2011 {published data only}
-
- Miron P. Prenatal screening for trisomy 21 and other aneuploidies at first trimester [Dépistage prénatal de la trisomie 21 et autres aneuploïdies au premier trimestre]. Université de Montréal (Faculté de Médecine) 2011:244 pages. [No AMICUS: 39273224]
Mundy 2008 {published data only}
-
- Mundy L, Hiller JE. Non‐invasive prenatal diagnostic test for Down's Syndrome (Structured abstract). Health Technology Assessment Database. Adelaide; Adelaide Health Technology Assessment (AHTA), 2008, issue 2. [HTA‐32012000229]
Mundy 2009 {published data only}
-
- Mundy L, Hiller JE. Non‐invasive prenatal diagnostic test for trisomy‐21 (Down's Syndrome) (Structured abstract). Health Technology Assessment Database. Adelaide Health Technology Assessment (AHTA) on behalf of National Horizon Scanning Unit (HealthPACT and MSAC), 2009, issue 2. [HTA‐32010000766]
Musci 2014 {published data only}
-
- Musci T. The largest study on NIPT thus far: The next study. Journal of Maternal‐fetal & Neonatal Medicine 2014;27:16.
Musci 2014a {published data only}
-
- Musci T, Struble C, Wang E, Hooks J, Syngelaki A, Mar Gil M, et al. Risk assessment for fetal sex chromosome aneuploidies using digital analysis of selected regions (DANSRTM) assays. Prenatal Diagnosis 2014;34:1‐2.
NCT00770458 {unpublished data only}
-
- NCT00770458. Non‐invasive screening for fetal aneuploidy: a new maternal plasma marker. ClinicalTrials.gov/show/NCT00770458 Date first received: 8 October 2008.
NCT00877292 {unpublished data only}
-
- NCT00877292. A new prenatal blood test for Down syndrome. ClinicalTrials.gov/show/NCT00877292 Date first received: 6 April 2009.
NCT00891852 {unpublished data only}
-
- NCT00891852. Non‐invasive determination of fetal chromosome abnormalities. ClinicalTrials.gov/show/NCT00891852 Date first received: 29 April 2009.
NCT00971334 {unpublished data only}
-
- NCT00971334. Noninvasive screening for fetal aneuploidy: a new maternal plasma marker. ClinicalTrials.gov/show/NCT00971334 Date first received: 1 September 2009.
NCT01052688 {unpublished data only}
-
- NCT01052688. Noninvasive screening for affected pregnancies: assay development and optimization in affected pregnancies. ClinicalTrials.gov/show/NCT01052688 Date first received: 15 January 2010.
NCT01256606 {unpublished data only}
-
- NCT01256606. Prenatal test for fetal aneuploidy detection. ClinicalTrials.gov/show/NCT01256606 Date first received: 22 November 2010.
NCT01451671 {unpublished data only}
-
- NCT01451671. Development of a prenatal test for fetal aneuploidy detection. ClinicalTrials.gov/show/NCT01451671 Date first received: 10 October 2011.
NCT01451684 {unpublished data only}
-
- NCT01451684. Development of a non‐invasive prenatal test. ClinicalTrials.gov/show/NCT01451684 Date first received: 10 October 2011.
NCT01555346 {unpublished data only}
-
- NCT01555346. Clinical evaluation of the SEQureDx T21 test In high risk pregnancies. ClinicalTrials.gov/show/NCT01555346 Date first received: 15 February 2012.
NCT01574781 {unpublished data only}
-
- NCT01574781. Non‐invasive prenatal diagnostic validation study. ClinicalTrials.gov/show/NCT01574781 Date first received: 2 April 2012.
NCT01597063 {unpublished data only}
-
- NCT01597063. Clinical evaluation of the SEQureDx Trisomy test in low risk pregnancies. ClinicalTrials.gov/show/NCT01597063 Date first received: 9 May 2012.
NCT01661010 {unpublished data only}
-
- NCT01661010. The clinical study of sex chromosome variants. ClinicalTrials.gov/show/NCT01661010 Date first received: 7 August 2012.
NCT01663675 {unpublished data only}
-
- NCT01663675. Trisomy 21 in adulthood. ClinicalTrials.gov/show/NCT01663675 Date first received: 8 August 2012.
NCT01668251 {unpublished data only}
-
- NCT01668251. Turner syndrome prenatal diagnosis study. ClinicalTrials.gov/show/NCT01668251 Date first received: 3 November 2011.
NCT01725438 {unpublished data only}
-
- NCT01725438. Non invasive prenatal diagnosis of trisomy 21 by genetic analysis of circulating fetal cells. ClinicalTrials.gov/show/NCT01725438 Date first received: 8 November 2012.
NCT01837979 {unpublished data only}
-
- NCT01837979. Down syndrome screening based on dried blood spots and cell‐free fetal DNA. ClinicalTrials.gov/show/NCT01837979 Date first received: 14 April 2013.
NCT01966991 {unpublished data only}
-
- NCT01966991. Prenatal screening for Down syndrome with DNAFirst. ClinicalTrials.gov/show/NCT01966991 Date first received: 17 October 2013.
NCT02127515 {unpublished data only}
-
- NCT02127515. Non invasive prenatal testing of Down syndrome. ClinicalTrials.gov/show/NCT02127515 Date first received: 2 April 2014.
NCT02226315 {unpublished data only}
-
- NCT02226315. Clinical performance of the MaterniT21™ PLUS LDT in multiple gestation pregnancies. ClinicalTrials.gov/show/NCT02226315 Date first received: 20 August 2014.
NCT02872948 {unpublished data only}
-
- NCT02872948. Diagnosis accuracy of noninvasive screening by PCR Digital for Down syndrome. ClinicalTrials.gov/show/NCT02872948 Date first received: 16 August 2016.
Neufeld‐Kaiser 2015 {published data only}
Neveling 2015 {published data only}
-
- Neveling K, Thung DT, Beulen L, Buijsman W, Gomes I, Heuvel S, et al. Clinical utility of NextSeq 500 sequencing for noninvasive prenatal testing. Prenatal Diagnosis 2015;35:54‐5.
Nickolich 2016 {published data only}
Nicolaides 2013a {published data only}
-
- Nicolaides K, Syngelaki A, Ashoor G, Musci T, Wang E, Song K. Clinical performance comparison of Harmony™ Prenatal Test and first‐trimester combined screening in general pregnancy population. Prenatal Diagnosis 2013;33:3.
Nicolaides 2014 {published data only}
-
- Nicolaides KH, Syngelaki A, Poon LC, Gil MM, Wright D. First‐trimester contingent screening for trisomies 21, 18 and 13 by biomarkers and maternal blood cell‐free DNA testing. Fetal Ddiagnosis and Therapy 2014;35(3):185‐92. [PUBMED: 24192489] - PubMed
Nicolaides 2014b {published data only}
-
- Nicolaides KH, Musci TJ, Struble CA, Syngelaki A, Mar Gil M. Assessment of fetal sex chromosome aneuploidy using directed cell‐free DNA analysis. Obstetrical and Gynecological Survey 2014;69(5):249‐50. - PubMed
Nicolaides 2014c {published data only}
-
- Nicolaides KH, Syngelaki A, Mar Gil M, Quezada MS, Zinevich Y. Prenatal detection of fetal triploidy from cell‐free DNA testing in maternal blood. Fetal Diagnosis and Therapy 2014;35(3):212‐7. [PUBMED: 24135152] - PubMed
Norem 2015 {published data only}
-
- Norem C, Obolensky E, Bijesse E, Turocy J, Blumberg B, Fehlen‐Quizon P, et al. Non‐invasive prenatal screening for trisomies‐2 years experience in a large Health Maintenance Organization (HMO). Prenatal Diagnosis 2015;35:62.
Norton 2014 {published data only}
-
- Norton ME, Jelliffe‐Pawlowski LL, Currier RJ. Chromosome abnormalities detected by current prenatal screening and noninvasive prenatal testing. Obstetrics and Gynecology 2014;124(5):979‐86. - PubMed
Norton 2014a {published data only}
-
- Norton ME, Jacobsson B, Swamy G, Laurent L, Ranzini A, Brar H, et al. Non‐invasive EXamination of Trisomy using directed cell‐free DNA analysis: The NEXT study. Prenatal Diagnosis 2014;34:e2.
Norton 2015a {published data only}
-
- Norton M, Wapner R, Kuppermann M, Jelliffe‐Pawlowski L, Currier R. Cell free DNA analysis vs sequential screening as primary testing considering all fetal chromosomal abnormalities. American Journal of Obstetrics and Gynecology 2015;212(1):S2. - PubMed
Norton 2015b {published data only}
Norton 2015c {published data only}
-
- Norton ME, Wapner RJ. Cell‐free DNA analysis for noninvasive examination of trisomy. New England Journal of Medicine 2015; Vol. 373, issue 26:2582. [PUBMED: 26699179] - PubMed
Norton 2016 {published data only}
-
- Norton ME, Baer RJ, Wapner RJ, Kuppermann M, Jelliffe‐Pawlowski LL, Currier RJ. Cell‐free DNA vs sequential screening for the detection of fetal chromosomal abnormalities. American Journal of Obstetrics and Gynecology 2016;214(6):727.e1‐6. [PUBMED: 26709085] - PubMed
O'Leary 2014 {published data only}
-
- O'Leary P, Maxwell S, Murch A, Dickinson J. What could non‐invasive prenatal testing miss in highrisk pregnancies and would it change outcomes?. Prenatal Diagnosis 2014;34:52‐3.
Oepkes 2015 {published data only}
-
- Oepkes D, Schuring‐Blom H, Pajkrt E, Faas B, Bax C, Coumans A, et al. TRIDENT: Or monitored NIPT implementation in the Netherlands. Prenatal Diagnosis 2015;35:10.
Oneda 2016 {published data only}
-
- Oneda B, Steindl K, Masood R, Reshetnikova I, Krejci P, Baldinger R, et al. Noninvasive prenatal testing: More caution in counseling is needed in high risk pregnancies with ultrasound abnormalities. European Journal of Obstetrics Gynecology and Reproductive Biology 2016;200:72‐5. - PubMed
Ordoñez 2015 {published data only}
-
- Ordoñez E, Rueda LR, Cañadas MP, Moreno M, Palao B, Cirigliano V. cfDNA‐Based aneuploidy screening, the more the better?. Prenatal Diagnosis 2015;35:107.
Palomaki 2011 {published data only}
-
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genetics in Medicine 2011;13(11):913‐20. [PUBMED: 22005709] - PubMed
Palomaki 2012a {published data only}
-
- Palomaki GE, Kloza EM, Canick JA. Evidence that next‐generation sequencing of maternal plasma will identify aneuploidies in a low risk population. Prenatal Diagnosis 2012;32:4.
Palomaki 2012b {published data only}
-
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. Editorial comment on DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Obstetrical and Gynecological Survey 2012;67(2):86‐8. [ISSN 0029‐7828] - PubMed
Palomaki 2015 {published data only}
-
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Boom D, Ehrich M, Deciu C, et al. Circulating cell free DNA testing: Are some test failures informative?. Prenatal Diagnosis 2015;35(3):289‐93. - PubMed
Palomaki 2015a {published data only}
-
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Boom D, Ehric M, Deciu C, et al. Circulating cell free DNA testing: are some test failures informative?. Obstetrical and Gynecological Survey 2015;70(8):492‐4. - PubMed
Perez‐Pedregosa 2015 {published data only}
-
- Pérez‐Pedregosa J, Paredes Ros B, Calles Hernandez LC, Izquierdo López L, Cabrillo Rodriguez E, Hurtado Caballero IV, et al. Non‐invasive prenatal screening for aneuploidy through analysis of cell‐free fetal DNA from maternal blood [Cribado prenatal no invasivo de aneuploidías mediante análisis de ADN fetal en sangre materna]. Progresos de Obstetricia y Ginecología 2015;58(3):113‐7.
Pescia 2017 {published data only}
Petersen 2014 {published data only}
-
- Petersen OB, Vogel I, Ekelund C, Hyett J, Tabor A. Potential diagnostic consequences of applying non‐invasive prenatal testing: population‐based study from a country with existing first‐trimester screening. Ultrasound in Obstetrics & Gynecology 2014;43(3):265‐71. - PubMed
Pettit 2014 {published data only}
-
- Pettit KE, Hull AD, Korty L, Jones MC, Pretorius DH. The utilization of circulating cell‐free fetal DNA testing and decrease in invasive diagnostic procedures: an institutional experience. Journal of Perinatology 2014;34(10):750‐3. [PUBMED: 24875410] - PubMed
Porreco 2014a {published data only}
-
- Porreco RP. Reply. American Journal of Obstetrics and Gynecology 2014;211(6):712. - PubMed
Rabinowitz 2012 {published data only}
Rabinowitz 2012a {published data only}
Rabinowitz 2012b {published data only}
-
- Rabinowitz M, Gemelos G, Banjevic M, Zimmermann B, Baner J, Levy B, et al. Highly accurate first trimester non‐invasive detection of fetal aneuploidy of 13, 18, 21 and sex chromosomes by targeted sequencing. Human Reproduction 2012;27(Suppl 2):ii106‐ii108. [ISSN 0268‐1161]
Rabinowitz 2013 {published data only}
-
- Rabinowitz M, Hill M, Demko Z, McAdoo S, Zimmermann B, Levy B. Using targeted sequencing of SNPs to achieve a highly accurate non‐invasive detection of fetal aneuploidy of 13,18, 21 and sex chromosomes. American Journal of Obstetrics and Gynecology 2013;208(1):S253.
Rabinowitz 2014 {published data only}
-
- Rabinowitz M, Valenti E, Pettersen B, Sigurjonsson S, Hill M, Zimmermann B. Noninvasive aneuploidy detection by multiplexed amplification and sequencing of polymorphic loci. Obstetrics and Gynecology 2014;123 Suppl 1(5):167S‐S.
Rad 2014 {published data only}
-
- Rad S, Turner AL, Beauchamp S, Aghajanian P, Williams J 3rd, Esakoff TF. Noninvasive prenatal testing compared with invasive diagnostic testing in the setting of an abnormal state aneuploidy screen. Obstetrics and Gynecology 2014;123 Suppl 1:100s.
Radoi 2015 {published data only}
Rava 2012 {published data only}
-
- Rava R, Bianchi D, Platt L, Goldberg J, Abuhamad A, Sehnert A. Genome wide fetal aneuploidy detection by sequencing of maternal plasma DNA: diagnostic accuracy in a prospective, blinded, multicenter study. American Journal of Obstetrics and Gynecology 2012;206(1):S367‐S.
Rava 2014 {published data only}
-
- Rava RP, Srinivasan A, Sehnert AJ, Bianchi DW. Circulating fetal cell‐free DNA fractions differ in autosomal aneuploidies and monosomy X. Clinical Chemistry 2014;60(1):243‐50. - PubMed
Reiff 2015 {published data only}
-
- Reiff E, Little S, Dobson L, Agarwala V, Wilkins‐Haug L, Bromley B. The role of first trimester ultrasound in prenatal aneuploidy screening for women with a negative cell free fetal DNA. Prenatal Diagnosis 2015;35:16‐7.
Reiff 2016 {published data only}
-
- Reiff ES, Little SE, Dobson L, Wilkins‐Haug L, Bromley B. What is the role of the 11‐ to 14‐week ultrasound in women with negative cell‐free DNA screening for aneuploidy?. Prenatal Diagnosis 2016;36(3):260‐5. [PUBMED: 26748490] - PubMed
Reimers 2015 {published data only}
-
- Reimers R, Mason‐Suares H, Little S, Reiff E, Bromley B, Dobson L, et al. What will be missed if Noninvasive Prenatal Testing (NIPT) is the only assessment for “Ultrasound Indicated” genetic studies?. Prenatal Diagnosis 2015;35:63‐4.
Revello 2016 {published data only}
-
- Revello R, Sarno L, Ispas A, Akolekar R, Nicolaides KH. Screening for trisomies by cell‐free DNA testing of maternal blood: consequences of a failed result. Ultrasound in Obstetrics & Gynecology 2016;47(6):698‐704. [PUBMED: 26743020] - PubMed
Ryan 2016 {published data only}
-
- Ryan A, Hunkapiller N, Banjevic M, Vankayalapati N, Fong N, Jinnett KN, et al. Validation of an enhanced version of a single‐nucleotide polymorphism‐based noninvasive prenatal test for detection of fetal aneuploidies. Fetal Diagnosis and Therapy 2016;40:219‐23. [PUBMED: 27028530] - PubMed
Sachse 2015 {published data only}
-
- Sachse M, Grömminger S, Schöck U, Bonnet J, Hofmann W. Development of a non‐invasive prenatal test (NIPT) assay for trisomy 21 (T21) based on comparative quantification of chromosome 21 and a reference chromosome via quantitative real‐time PCR. Medizinische Genetik 2015;27(1):177.
Samura 2015 {published data only}
-
- Samura O, Sago H, Sekizawa A. Nationwide project using next‐generation sequencing of cellfree DNA in maternal plasma in Japan: One and a half years of experience. Prenatal Diagnosis 2015;35:64. - PubMed
Sarno 2016 {published data only}
-
- Sarno L, Revello R, Hanson E, Akolekar R, Nicolaides KH. Prospective first‐trimester screening for trisomies by cell‐free DNA testing of maternal blood in twin pregnancy. Ultrasound in Obstetrics & Gynecology 2016;47(6):705‐11. [PUBMED: 26970114] - PubMed
Schöck 2015 {published data only}
-
- Schöck U, Grömminger S, Bonnet J, Sachse M, Hofmann W. Recent bioinformatic advances of non‐invasive prenatal detection to enhance diagnostic accuracy and aneuploidy discovery. Medizinische Genetik 2015;27(1):177.
Sehnert 2013 {published data only}
Sehnert 2014 {published data only}
-
- Sehnert A, Bianchi D, Rava R. Maternal cell‐free DNA (cfDNA) sequencing versus standard prenatal aneuploidy screening in a general obstetrical population. Prenatal Diagnosis 2014;34:8.
Sentilhes 2015 {published data only}
-
- Sentilhes L, Salomon LJ, Vayssiere C. Cell‐free DNA analysis for noninvasive examination of trisomy. New England Journal of Medicine 2015; Vol. 373, issue 26:2581‐2. [PUBMED: 26699181] - PubMed
Seo 2015 {published data only}
-
- Seo DH, Cho DY, Kim J, Kim SY, Cho SE, Oh M. Clinical study of non‐invasive prenatal testing using next‐generation sequencing. Journal of Laboratory Medicine and Quality Assurance 2015;37:214‐8. [DOI: 10.15263/jlmqa.2015.37.4.214] - DOI
Settler 2015 {published data only}
-
- Settler C, Dohany L. Clinical laboratory experience in a general obstetrical population of noninvasive prenatal screening for fetal aneuploidy from cell free DNA. Prenatal Diagnosis 2015;35:64‐5.
Shani 2016 {published data only}
-
- Shani H, Goldwaser T, Keating J, Klugman S. Chromosomal abnormalities not currently detected by cell‐free fetal DNA: a retrospective analysis at a single center. American Journal of Obstetrics and Gynecology 2016;214(6):729.e1‐729.e11. [PUBMED: 26721783] - PubMed
Shaohua 2012 {published data only}
-
- Shaohua T, Huanzheng L, Erle Z, Xueqing X. Noninvasive prenatal diagnosis for aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Prenatal Diagnosis 2012;32:79.
Sharma 2015 {published data only}
-
- Sharma P, Metcalfe A, Pastuck M, Laberge A‐M, Haidar H, Ravitsky V, et al. Women's understanding of non‐invasive prenatal testing based on cell free DNA versus first trimester combined screening. Prenatal Diagnosis 2015;35:106.
Shaw 2013 {published data only}
-
- Shaw SWS, Chen CY, Hsiao CH, Ren Y, Tian F, Tsai C, et al. Non‐invasive prenatal testing for whole fetal chromosomal aneuploidies: A multi‐center prospective cohort trial in Taiwan. Prenatal Diagnosis 2013;33:81. - PubMed
Shen 2016 {published data only}
-
- Shen J, Wen Z, Qin X, Shi Y. Noninvasive fetal trisomy detection by multiplexed semiconductor sequencing: a barcoding analysis strategy. Journal of Human Genetics 2016;61(3):247‐52. [PUBMED: 26657936] - PubMed
Shi 2015 {published data only}
-
- Shi X, Zhang Z, Cram DS, Liu C. Feasibility of noninvasive prenatal testing for common fetal aneuploidies in an early gestational window. Clinica Chimica Acta 2015;439:24‐8. - PubMed
Shulman 2014 {published data only}
-
- Shulman L, Dungan J, Ginsberg N. The use of noninvasive prenatal screening (NIPS) in the assessment of an abnormal fetal ultrasound. Prenatal Diagnosis 2014;34:62.
Sistermans 2015a {published data only}
-
- Sistermans E, Straver R, Faas BH. Maternal malignancies detected with noninvasive prenatal testing. JAMA 2015; Vol. 314, issue 20:2192. [PUBMED: 26599191] - PubMed
Smith‐Bindman 2015 {published data only}
-
- Smith‐Bindman R, Miglioretti D. Cell‐free DNA analysis for noninvasive examination of trisomy. New England Journal of Medicine 2015; Vol. 373, issue 26:2581. [PUBMED: 26699180] - PubMed
Song 2012 {published data only}
-
- Song K, Ashoor G, Syngelaki A, Wagner M, Birdir C, Struble C, et al. Clinical evaluation of a directed cfDNA analysis method for non‐invasive prenatal fetal trisomy detection. Prenatal Diagnosis 2012;32:16‐7.
Sparks 2012 {published data only}
Srinivasan 2013 {published data only}
-
- Srinivasan A, Bianchi D, Liao W, Sehnert A, Rava R. Maternal plasma DNA sequencing: Effects of multiple gestation on aneuploidy detection and the relative cell‐free fetal DNA (cffDNA) per fetus. American Journal of Obstetrics and Gynecology 2013;208(1):S31.
Stokowski 2015 {published data only}
-
- Stokowski R, Wang E, White K, Batey A, Jacobsson B, Brar H, et al. Clinical performance of non‐invasive prenatal testing (NIPT) using targeted cell‐free DNA analysis in maternal plasma with microarrays or next generation sequencing (NGS) is consistent across multiple controlled clinical studies. Prenatal Diagnosis 2015;35(12):1243‐6. [PUBMED: 26332378] - PMC - PubMed
Strah 2015 {published data only}
-
- Strah D, Ovniček P, Bernik J. Non‐invasive prenatal cell‐free fetal DNA testing for down syndrome and other chromosomal abnormalities [Neinvazivno predrojstveno testiranje prostih plodov DNA za downov sindrom in ostale kromosomske nepravilnosti]. Zdravniški Vestnik 2015;84(11):727‐33.
Straver 2014 {published data only}
Strom 2015 {published data only}
-
- Strom C. The positive predictive value of noninvasive prenatal screening (NIPS). Chromosome Research 2015;23(1):S23.
Stumm 2011 {published data only}
-
- Stumm M, Trunk N, Beck M, Entezami M, Becker R, Locherbach J, et al. Non‐invasive prenatal detection of chromosome aneuploidies using next‐generation sequencing: First steps towards clinical application. Medizinische Genetik 2011;23(1):86.
Stumm 2012 {published data only}
-
- Stumm M, Entezami M, Trunk N, Beck M, Locherbach J, Wegner RD, et al. Noninvasive prenatal detection of chromosomal aneuploidies using different next generation sequencing strategies and algorithms. Prenatal Diagnosis 2012;32(6):569‐77. [PUBMED: 22573401] - PubMed
Stumm 2012a {published data only}
-
- Stumm M, Entezami M, Haug K, Blank C, Wustemann M, Schulze B, et al. Non‐invasive prenatal detection of trisomy 21 using massively parallel sequencing: A collaborative study in Europe. Prenatal Diagnosis 2012;32:63‐4. - PubMed
Stumm 2013 {published data only}
-
- Stumm M, Entezami M, Haug K, Blank C, Wustemann C, Schulze B, et al. Diagnostic accuracy of PrenaTest® for non‐invasive prenatal detection of common autosomal aneuploidies. Medizinische Genetik 2013;25(1):171‐2. - PubMed
Stumm 2016 {published data only}
-
- Stumm M. Noninvasive prenatal tests from the viewpoint of human geneticists: Possibilities and limits in daily routine [Nichtinvasive pränatale Tests aus Sicht des Humangenetikers: Möglichkeiten und Grenzen in der täglichen Routine]. Der Gynäkologe 2016;49(6):429‐36.
Swanson 2012 {published data only}
-
- Swanson A, Coffeen C, Sehnert AJ. Non‐invasive prenatal testing for fetal aneuploidy by massively parallel DNA sequencing of maternal plasma: The future has arrived today. Laboratoriums Medizin 2012;36(5):269‐75.
Syngelaki 2014 {published data only}
-
- Syngelaki A, Pergament E, Homfray T, Akolekar R, Nicolaides KH. Replacing the combined test by cell‐free DNA testing in screening for trisomies 21, 18 and 13: impact on the diagnosis of other chromosomal abnormalities. Fetal Diagnosis and therapy 2014;35(3):174‐84. [PUBMED: 24525399] - PubMed
Tan 2016 {published data only}
-
- Tan Y, Gao Y, Lin G, Fu M, Li X, Yin X, et al. Noninvasive prenatal testing (NIPT) in twin pregnancies with treatment of assisted reproductive techniques (ART) in a single center. Prenatal Diagnosis 2016;36(7):672‐9. [PUBMED: 27150972] - PubMed
Taneja 2016 {published data only}
Taneja 2017 {published data only}
-
- Taneja PA, Prosen TL, Feo E, Kruglyak KM, Halks‐Miller M, Curnow KJ, et al. Fetal aneuploidy screening with cell‐free DNA in late gestation. Journal of Maternal‐Fetal & Neonatal Medicine 2017;30(3):338‐42. [PUBMED: 27124739] - PubMed
Tarrier 2015 {published data only}
-
- Tarrier B, Sekedat M, Mann T, Stoerker J, Bombard A. Whole genome maternal plasma DNA sequencing for fetal aneuploidy accurately detects autosomal and sex chromosome abnormalities: A validation study with independent fetal fraction observations. Prenatal Diagnosis 2015;35:65‐6.
Taylor 2014 {published data only}
-
- Taylor JB, Chock VY, Hudgins L. NIPT in a clinical setting: an analysis of uptake in the first months of clinical availability. Journal of Genetic Counseling 2014;23(1):72‐8. - PubMed
Togneri 2016 {published data only}
-
- Togneri F, Court S, Parks M, Clokie S, Hamilton S, Bibb N, et al. Noninvasive prenatal testing for fetal aneuploidy: The experience of an NHS regional genetics laboratory. BJOG: an international journal of obstetrics and gynaecology 2016;123:70.
Tong 2016 {published data only}
-
- Tong H, Jin Y, Xu Y, Zou B, Ye H, Wu H, et al. Prenatal diagnosis of trisomy 21, 18 and 13 by quantitative pyrosequencing of segmental duplications. Clinical Genetics 2016;90(5):451‐5. [PUBMED: 26948280] - PubMed
Valderramos 2016a {published data only}
-
- Valderramos SG, Rao R, Scibetta EW, Clark K, Krakow D, Silverman NS, et al. Clinical accuracy of abnormal cell‐free fetal DNA (cfDNA) results for the sex chromosomes. American Journal of Obstetrics and Gynecology 2016;214(1):S402‐3.
Valderramos 2016b {published data only}
Valderramos 2016c {published data only}
van den Oever 2012a {published data only}
-
- Oever JM, Balkassmi S, Verweij EJ, Iterson M, Adama van Scheltema PN, Oepkes D, et al. Single molecule sequencing of free DNA from maternal plasma for noninvasive trisomy 21 detection. Clinical Chemistry 2012;58(4):699‐706. [PUBMED: 22278607] - PubMed
van den Oever 2012b {published data only}
-
- Oever JME, Balkassmi S, Verweij EJ, Iterson M, Scheltema PNA, Oepkes D, et al. Single molecule sequencing of free DNA from maternal plasma for non‐invasive trisomy 21 detection. Prenatal Diagnosis 2012;32:60‐1.
van den Oever 2013 {published data only}
-
- Oever JM, Balkassmi S, Johansson LF, Adama van Scheltema PN, Suijkerbuijk RF, Hoffer MJ, et al. Successful noninvasive trisomy 18 detection using single molecule sequencing. Clinical Chemistry 2013;59(4):705‐9. [PUBMED: 23315481] - PubMed
Van Opstal 2016 {published data only}
Verweij 2013a {published data only}
-
- Verweij EJ, Jacobsson B, Scheltema PNA, Boer MA, Hoffer MJV, Hollemon D, et al. European Noninvasive Trisomy Evaluation (EU‐NITE) study: Multicenter prospective study for noninvasive fetal trisomy 21 testing. Prenatal Diagnosis 2013;33:83‐4. - PubMed
Wald 2015a {published data only}
-
- Wald NJ, Bestwick JP. Performance of antenatal reflex DNA screening for Down's syndrome. Journal of Medical Screening 2015;22(4):168‐74. [PUBMED: 25882667] - PubMed
Wald 2015b {published data only}
-
- Wald NJ, Huttly WJ, Bestwick JP, Aquilina J, Peregrine E. Reflex antenatal DNA screening for Down syndrome. Prenatal Diagnosis 2015; Vol. 35, issue 11:1154. [PUBMED: 26211510] - PubMed
Wang 2012 {published data only}
-
- Wang SJ, Gao ZY, Lu YP, Li YL, You YQ, Zhang LW, et al. Value of detection of cell‐free fetal DNA in maternal plasma in the prenatal diagnosis of chromosomal abnormalities. Zhonghua Fu Chan Ke za Zhi 2012;47(11):808‐12. [PUBMED: 23302119] - PubMed
Wang 2015b {published data only}
-
- Wang JC, Sahoo T, Schonberg S, Kopita KA, Ross L, Patek K, et al. Discordant noninvasive prenatal testing and cytogenetic results: a study of 109 consecutive cases. Genetics in Medicine 2015;17(3):234‐6. [PUBMED: 25101914] - PubMed
Wang 2015c {published data only}
-
- Wang S, Huang S, Ma L, Liang L, Zhang J, Zhang J, et al. Maternal X chromosome copy number variations are associated with discordant fetal sex chromosome aneuploidies detected by noninvasive prenatal testing. Clinica Chimica Acta; International Journal of Clinical Chemistry 2015;444:113‐6. [PUBMED: 25689220] - PubMed
Wang 2015d {published data only}
-
- Wang JC, Sahoo T, Schonberg S, Kopita KA, Ross L, Patek K, et al. Discordant noninvasive prenatal testing and cytogenetic results: A study of 109 consecutive cases. Obstetrical and Gynecological Survey 2015;70(7):434‐6. - PubMed
Wang 2015e {published data only}
-
- Wang E, Struble C, Kingsley C, Steeke R, Batey A, Hollemon D, et al. Importance of fetal fraction analysis for CFDNA testing in the general pregnancy population. Reproductive Sciences 2015;22:176A.
Xiong 2015 {published data only}
-
- Xiong Y, Berman J, Weiner S, Carre A, Seligman NS, Steinbach G, et al. Noninvasive prenatal testing (NIPT) registry‐patients' results and providers' perspective. Reproductive Sciences 2015;22:176A‐7A.
Yankova 2015 {published data only}
-
- Yankova M, Chaveeva P, Stratieva V. Models of clinical implementation of cell free fetal DNA in the maternal serum screening test‐analysis. Akusherstvo i Ginekologiia 2015;54(7):15‐21. [PUBMED: 27025103] - PubMed
Yaron 2015 {published data only}
-
- Yaron Y, Jani J, Schmid M, Oepkes D. Current status of testing for microdeletion syndromes and rare autosomal trisomies using cell‐free DNA technology. Obstetrics and Gynecology 2015;126(5):1095‐9. [PUBMED: 26444108] - PubMed
Yeang 2014 {published data only}
-
- Yeang CH, Ma GC, Hsu HW, Lin YS, Chang SM, Cheng PJ, et al. Genome‐wide normalized score: a novel algorithm to detect fetal trisomy 21 during non‐invasive prenatal testing. Ultrasound in Obstetrics & Gynecology 2014;44(1):25‐30. [PUBMED: 24700679] - PubMed
Yu 2014 {published data only}
Yuan 2013 {published data only}
-
- Yuan Y, Jiang F, Hua S, Du B, Hao Y, Ye L, et al. Feasibility study of semiconductor sequencing for noninvasive prenatal detection of fetal aneuploidy. Clinical Chemistry 2013;59(5):846‐9. [PUBMED: 23364181] - PubMed
Zhang 2015 {published data only}
-
- Zhang H, Gao Y, Jiang F, Fu M, Yuan Y, Guo Y, et al. Non‐invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound in Obstetrics & Gynecology 2015;45(5):530‐8. [PUBMED: 25598039] - PubMed
Zhou 2013 {published data only}
-
- Zhou D, Liang D, Lv W, Tian F, Song Z, Zhang J, et al. Noninvasive prenatal testing in China: Results and learning from a clinical study of more than 50,000 pregnancies. Prenatal Diagnosis 2013;33:1.
Zimmermann 2012 {published data only}
Zimmermann 2013 {published data only}
-
- Zimmermann B, Banjevic M, Hill M, Lacroute P, Dodd M, Sigurjonsson S, et al. Highly multiplexed single‐nucleotide polymorphism amplification and sequencing to identify fetal aneuploidy from maternal cell‐free DNA. Prenatal Diagnosis 2013;33:68.
Zwiefelhofer 2013 {published data only}
-
- Zwiefelhofer T, Whitley P, Roy K, Jean‐Jacques R, Ehrich M. Prenatal detection of fetal aneuploidy using bench top sequencing. Prenatal Diagnosis 2013;33:85.
References to ongoing studies
Basaran 2015 {published and unpublished data}
-
- Basaran S, Yuksel A, Has R, Kirgiz M, Dehgan T, Karaman B. False positive and false negative results of cell free DNA testing. Chromosome Research 2015;23(1):S124‐S5.
Buresch 2016 {published data only}
-
- Buresch A, Rosner M, Suskin B, Einstein F, Bircaj E, Lister R, et al. Actual rates of recommended diagnostic testing after first trimester screening vs. same‐day screening by cell free DNA. American Journal of Obstetrics and Gynecology 2016;214(1):S326.
Chen 2011a {published data only}
-
- Chen F, Wang W, Shan D, Ren J, Xie J, Huang Y, et al. Noninvasive prenatal diagnosis of fetal aneuploidy by massively parallel sequencing of maternal plasma DNA. Journal of Perinatal Medicine 2011;39:299.
Da Fonseca 2015 {published data only}
-
- Fonseca JP, Khattabi L, Brun S, Chatron N, Gueguen P, Nectoux J, et al. Non‐Invasive prenatal testing for the most common aneuploidies (trisomies 21, 18, and 13) using a semiconductor‐sequencing platform: a French multicenter pilot study. Chromosome Research 2015;23(1):S110‐S1.
ISRCTN11174071 {unpublished data only}
-
- ISRCTN11174071. Comparison of false positive rates in prenatal combined screening and cell free DNA screening for trisomy 21. isrctn.com/ISRCTN11174071 Date first received: 26 July 2016. [DOI: 10.1186/ISRCTN11174071] - DOI
Lin 2014 {published data only}
-
- Lin G, Gao Y, Yin X, Tan Y, Zhang H, Lu G, et al. Clinical implementation of noninvasive prenatal testing in twin pregnancies with assisted reproductive technique treatment. Prenatal Diagnosis 2014;34:14.
Mu 2014 {published data only}
-
- Mu Y, Sun F. Maternal non‐invasive fetal DNA test used in prenatal diagnosis. International Journal of Laboratory Hematology 2014;36:106‐7.
NCT01429389 {unpublished data only}
-
- NCT01429389. Specimen collection from pregnant women at increased risk for fetal aneuploidy. ClinicalTrials.gov/show/NCT01429389 Date first received: 2 September 2011.
NCT01472523 {unpublished data only}
-
- NCT01472523. A safer pre‐natal diagnosis using free DNA in maternal blood. ClinicalTrials.gov/show/NCT01472523 Date first received: 11 October 2011.
NCT01545674 {unpublished data only}
-
- NCT01545674. Prenatal non‐invasive aneuploidy test utilizing SNPs trial. ClinicalTrials.gov/show/NCT01545674 Date first received: 1 March 2012.
NCT01925742 {unpublished data only}
-
- NCT01925742. Study of the efficacy of new non‐invasive prenatal tests for screening for fetal trisomies using maternal blood. ClinicalTrials.gov/show/NCT01925742 Date first received: 16 August 2013.
NCT02201862 {unpublished data only}
-
- NCT02201862. Non‐invasive chromosomal evaluation of trisomy study. ClinicalTrials.gov/show/NCT02201862 Date first received: 22 July 2014.
NCT02278536 {unpublished data only}
-
- NCT02278536. Multiple gestation study. ClinicalTrials.gov/show/NCT02278536 Date first received: 28 October 2014.
NCT02278874 {unpublished data only}
-
- NCT02278874. High risk multiple gestation study. ClinicalTrials.gov/show/NCT02278874 Date first received: 27 August 2014.
NCT02317965 {unpublished data only}
-
- NCT02317965. Non‐invasive screening for fetal aneuploidy. ClinicalTrials.gov/show/NCT02317965 Date first received: 9 December 2014.
NCT02424474 {unpublished data only}
-
- NCT02424474. T21,18 and 13 screening by cell free fetal DNA in low risk patients. ClinicalTrials.gov/show/NCT02424474 Date first received: 8 April 2015.
NCT02787486 {unpublished data only}
-
- NCT02787486. Expanded noninvasive genomic medical assessment: the Enigma Study. ClinicalTrials.gov/show/NCT02787486 Date first received: 26 May 2016.
Sago 2015 {published data only}
-
- Sago H, Sekizawa A, and Japan NIPT consortium. Nationwide demonstration project of next‐generation sequencing of cell‐free DNA in maternal plasma in Japan: 1‐year experience. Prenatal Diagnosis 2015;35(4):331‐6. - PubMed
Sanchez‐Usabiaga 2015 {published data only}
-
- Sanchez‐Usabiaga RA, Aguinaga‐Rios M, Batista‐Espinoza A, Hurtado‐Amador R, Romero‐Tovar S. [Clinical implementation of non‐invasive prenatal study for detecting aneuploidies by fetal DNA based on single nucleotide polymorphisms: two years in Mexico] [Implementacion clinica del estudio prenatal no invasivo para Ia deteccion de aneuplodias mediante ADN fetal con base en polimorfismos de nucleotido unico: dos anos en Mexico]. Ginecologia y Obstetricia de Mexico 2015;83(4):220‐31. [PUBMED: 26727755] - PubMed
Sistermans 2015 {published data only}
-
- Sistermans EA, Schuring‐Blom GH, Faas BHW, Boon EMJ, Bax CJ, Coumans ABC, et al. TRIDENT: or monitored NIPT implementation in the Netherlands. European Journal of Human Genetics 2015;23(Suppl 1):C01.2.
Torres 2015 {published data only}
-
- Torres Y, Suela J, Cigudosa JC. Genetic non invasive prenatal testing: A clinical and technical experience of 3.000 cases with follow‐up. Chromosome Research 2015;23(1):S140‐S141. [ISSN 09673849]
Van Wymersch 2015 {published data only}
-
- Wymersch D, Gilson G. Introduction of noninvasive prenatal testing for fetal trisomies: preliminary results and consequences on invasive samplings [Introduction du depistage sanguin des trisomies foetales par recherche d'ADN foetal circulant: Resultats preliminaires d'une annee et evolution des prelevements invasifs]. Bulletin de la Societe des Sciences Medicales du Grand‐Duche de Luxembourg 2015;1:65‐72. [PUBMED: 26946853] - PubMed
Willems 2014 {published data only}
Yu 2014a {published data only}
-
- Yu M, Fei S. Maternal non‐invasive fetal DNA test used in prenatal diagnosis. Clinical Chemistry and Laboratory Medicine 2014;52:S1570.
Zwiefelhofer 2014 {published data only}
-
- Zwiefelhofer T, Whitley P, Roy K, Saha M, Burcham T, Boom D, et al. Prenatal detection of fetal aneuploidy on the Ion Torrent Proton™ platform. Prenatal Diagnosis 2014;34:64‐5.
Additional references
ACOG #163 2016
-
- American College of Obstetricians and Gynecologists. Practice Bulletin No. 163 Summary: Screening for fetal aneuploidy. Obstetrics and Gynecology 2016;127(5):979‐81. [PUBMED: 27101120] - PubMed
ACOG #545 2012
-
- American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 545: Noninvasive prenatal testing for fetal aneuploidy. Obstetrics and Gynecology 2012;120(6):1532‐4. [PUBMED: 23168792] - PubMed
ACOG #682 2016
-
- American College of Obstetricians and Gynecologists and Society for Maternal‐Fetal Medicine. Committee Opinion No.682: Microarrays and next‐generation sequencing technology: The use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstetrics and Gynecology 2016;128(6):e262‐8. [PUBMED: 27875474] - PubMed
ACOG #88 2007
-
- American College of Obstetricians and Gynecologists. ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstetrics and Gynecology 2007;110(6):1459‐67. [PUBMED: 18055749] - PubMed
Agarwal 2013
Akolekar 2015
-
- Akolekar R, Beta J, Picciarelli G, Ogilvie C, D'Antonio F. Procedure‐related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta‐analysis. Ultrasound in Obstetrics & Gynecology 2015;45(1):16‐26. [PUBMED: 25042845] - PubMed
Alldred 2010
-
- Alldred SK, Deeks JJ, Neilson JP, Alfirevic Z. Antenatal screening for Down's syndrome: generic protocol. Cochrane Database of Systematic Reviews 2010, Issue 4. [DOI: 10.1002/14651858.CD007384.pub2; CD007384] - DOI
Alldred 2012
Alldred 2015a
Alldred 2015b
Alldred 2017a
-
- Alldred SK, Takwoingi Y, Guo B, Pennant M, Deeks JJ, Neilson JP, et al. First trimester ultrasound tests alone or in combination with first trimester serum tests for Down's syndrome screening. Cochrane Database of Systematic Reviews 2017, Issue 3. [DOI: 10.1002/14651858.CD012600; PUBMED: 28295158] - DOI - PMC - PubMed
Alldred 2017b
-
- Alldred SK, Takwoingi Y, Guo B, Pennant M, Deeks JJ, Neilson JP, et al. First and second trimester serum tests with and without first trimester ultrasound tests for Down's syndrome screening. Cochrane Database of Systematic Reviews 2017, Issue 3. [DOI: 10.1002/14651858.CD012599; PUBMED: 28295159] - DOI - PMC - PubMed
APA 2013
-
- American Psychiatric Association. Neurodevelopmental disorders. Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DMS‐5®). Arlington: American Psychiatric Publishing, 2013. [DOI: 10.1176/appi.books.9780890425596.514988] - DOI
Ariosa Diagnostics 2016
-
- Ariosa Diagnostics, Inc, USA. www.ariosadx.fr/healthcare‐professionals/ (accessed 25 October 2016).
Attilakos 2011
-
- Attilakos G, Maddocks DG, Davies T, Hunt LP, Avent ND, Soothill PW, et al. Quantification of free fetal DNA in multiple pregnancies and relationship with chorionicity. Prenatal Diagnosis 2011;31(10):967‐72. [PUBMED: 21769896] - PubMed
Bardsley 2013
Basset 2013
-
- Basset C, Beaufils F, Dreifuss‐Netter F, Droit RP, Gaudray P, Matuchansky C, et al. Avis N° 120: Questions éthiques associées au développement des tests génétiques foetaux sur sang maternel. Comité Consultatif National d’Ethique (CCNE) pour les sciences de la vie et de la santé, 2013:50 pages.
Benn 2011
-
- Benn P, Borrell A, Crossley J, Cuckle H, Dugoff L, Gross S, et al. Aneuploidy screening: a position statement from a committee on behalf of the Board of the International Society for Prenatal Diagnosis, January 2011. Prenatal Diagnosis 2011;31(6):519‐22. [PUBMED: 21604286] - PubMed
Benn 2013a
-
- Benn P, Borell A, Chiu R, Cuckle H, Dugoff L, Faas B, et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenatal Diagnosis 2013;33(7):622‐9. [PUBMED: 23616385] - PubMed
Benn 2013b
-
- Benn P, Cuckle H, Pergament E. Non‐invasive prenatal testing for aneuploidy: current status and future prospects. Ultrasound in Obstetrics & Gynecology 2013;42(1):15‐33. [PUBMED: 23765643] - PubMed
Benn 2016
-
- Benn P. Expanding non‐invasive prenatal testing beyond chromosomes 21, 18, 13, X and Y. Clinical Genetics 2016;90(6):477‐85. [PUBMED: 27283893] - PubMed
Berry Genomics 2016
-
- Berry Genomics Co. Ltd, China. www.berrygenomics.com/bambnitest/ (accessed 5 December 2016).
BGI 2014
-
- Bejing Genomics Institute, China. www.niftytest.com/plus/list.php?tid=14 (accessed 20 June 2014).
BGI 2016
-
- Bejing Genomics Institute, China. www.niftytest.com/healthcare‐providers/clinical‐data/ (accessed 25 October 2016).
Bianchi 2004
-
- Bianchi DW. Circulating fetal DNA: its origin and diagnostic potential‐a review. Placenta 2004;25 Suppl A:S93‐S101. [PUBMED: 15033315] - PubMed
Birch 2005
-
- Birch L, English CA, O'Donoghue K, Barigye O, Fisk NM, Keer JT. Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clinical Chemistry 2005;51(2):312‐20. [PUBMED: 15608152] - PubMed
Bussani 2011
-
- Bussani C, Tommaso M, Cioni R, Pasquini L, Quitadamo L, Scarselli G. Quantitative variation analysis of fetal DNA in maternal plasma samples collected before and after amniocentesis. Journal of Obstetrics and Gynaecology Research 2011;37(6):571‐4. [PUBMED: 21375672] - PubMed
Canick 2013
-
- Canick JA, Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenatal Diagnosis 2013;33(7):667‐74. [PUBMED: 23592541] - PubMed
Cereda 2012
Chen 2009
-
- Chen CP. Prenatal sonographic features of fetuses in trisomy 13 pregnancies (I). Taiwanese Journal of Obstetrics & Gynecology 2009;48(3):210‐7. [PUBMED: 19797008] - PubMed
Chetty 2013
-
- Chetty S, Garabedian MJ, Norton ME. Uptake of noninvasive prenatal testing (NIPT) in women following positive aneuploidy screening. Prenatal Diagnosis 2013;33(6):542‐6. [PUBMED: 23592525] - PubMed
Chitayat 2011
-
- Chitayat D, Langlois S, Wilson RD, Genetics Committee of the Society of Obstetricians and Gynaecologists of Canada, Prenatal Diagnosis Committee of the Canadian College of Medical Geneticists. Prenatal screening for fetal aneuploidy in singleton pregnancies. Journal of Obstetrics and Gynaecology Canada : JOGC 2011;33(7):736‐50. [PUBMED: 21749752] - PubMed
Choi 2012
-
- Choi H, Riper M, Thoyre S. Decision making following a prenatal diagnosis of Down syndrome: an integrative review. Journal of Midwifery & Women's Health 2012;57(2):156‐64. [PUBMED: 22432488] - PubMed
Christianson 2006
-
- Christianson A, Howson CP, Modell B. March of dimes. Global report on birth defects. The hidden toll of dying and disabled children. March of Dimes Birth Defects Foundation 2006:1‐76. [http://www.marchofdimes.com/glue/files/global‐report‐on‐birth‐defects‐th...
Chu 2006
-
- Chu H, Cole SR. Bivariate meta‐analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach. Journal of Clinical Epidemiology 2006; Vol. 59, issue 12:1331‐2; author reply 1332‐3. [PUBMED: 17098577] - PubMed
Cochrane Glossary 2014
-
- The Cochrane Collaboration’s Glossary of terms. community‐archive.cochrane.org/glossary (accessed 17 July 2014).
Deeks 2013
-
- Deeks JJ, Wisniewski S, Davenport C. Chapter 4: Guide to the contents of a Cochrane Diagnostic Test Accuracy Protocol. In: Deeks JJ, Bossuyt PM, Gatsonis C (editors), Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2013. Available from: methods.cochrane.org/sdt/handbook‐dta‐reviews.
Devlin 2004
Driscoll 2009
-
- Driscoll DA, Gross S. Clinical practice. Prenatal screening for aneuploidy. New England Journal of Medicine 2009;360(24):2556‐62. [PUBMED: 19516035] - PubMed
Duncan 2011
-
- Duncan A, Langlois S. Use of array genomic hybridization technology in prenatal diagnosis in Canada. Journal of Obstetrics and Gynaecology Canada 2011;33(12):1256‐9. [PUBMED: 22166281] - PubMed
Fan 2010
-
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell‐free DNA by paired‐end sequencing. Clinical Chemistry 2010;56(8):1279‐86. [PUBMED: 20558635] - PubMed
Fishler 1991
-
- Fishler K, Koch R. Mental development in Down syndrome mosaicism. American Journal of Mental Retardation 1991;96(3):345‐51. [PUBMED: 1836734] - PubMed
Gagnon 2010
-
- Gagnon S, Labrecque M, Njoya M, Rousseau F, St‐Jacques S, Legare F. How much do family physicians involve pregnant women in decisions about prenatal screening for Down syndrome?. Prenatal Diagnosis 2010;30(2):115‐21. [PUBMED: 20013876] - PubMed
Gekas 2009
Genesupport 2016
-
- Genesupport, Switzerland. www.prendia.ch/fileadmin/user_upload/pdf/Tableau_Anomalies‐Numeriques.pdf (accessed 25 October 2016).
Genoma 2016
-
- Genoma, Switzerland. genoma.com/product_page.php?id=2 (accessed 10 November 2016).
Genome Care 2016
-
- Genome Care, Korea. http://genomecare.net/en/ (accessed 5 December 2016).
Gil 2015a
-
- Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell‐free DNA in maternal blood in screening for fetal aneuploidies: updated meta‐analysis. Ultrasound in Obstetrics & Gynecology 2015;45(3):249‐66. [PUBMED: 25639627] - PubMed
Groth 2013
-
- Groth KA, Skakkebaek A, Host C, Gravholt CH, Bojesen A. Clinical review: Klinefelter syndrome‐‐a clinical update. Journal of Clinical Endocrinology and Metabolism 2013;98(1):20‐30. [PUBMED: 23118429] - PubMed
HAS 2015
-
- Haute Autorité de Santé. Screening test accuracy for fetal trisomy 21 with circulating cell‐free DNA. [Les performances des tests de dépistage de la trisomie 21 foetale par analyse de l’ADN libre circulant]. Collège de la Haute Autorité de Santé 2015.
ICFMM 2013
Illumina 2014
-
- Verinata Health, Inc. www.verifitest.com/clinical‐data/ (accessed 3 March 2014).
Illumina 2016
-
- Illumina, Inc, USA. www.illumina.com/clinical/reproductive‐genetic‐health/nipt.html (accessed 25 October 2016).
Irving 2011
-
- Irving C, Richmond S, Wren C, Longster C, Embleton ND. Changes in fetal prevalence and outcome for trisomies 13 and 18: a population‐based study over 23 years. Journal of Maternal‐Fetal & Neonatal Medicine 2011;24(1):137‐41. [PUBMED: 20384468] - PubMed
Irving 2012
-
- Irving CA, Chaudhari MP. Cardiovascular abnormalities in Down's syndrome: spectrum, management and survival over 22 years. Archives of Disease in Childhood 2012;97(4):326‐30. [PUBMED: 21835834] - PubMed
Karnis 2012
-
- Karnis MF. Fertility, pregnancy, and medical management of Turner syndrome in the reproductive years. Fertility and Sterility 2012;98(4):787‐91. [PUBMED: 23020910] - PubMed
Kazerouni 2011
-
- Kazerouni NN, Currier RJ, Flessel M, Goldman S, Hennigan C, Hodgkinson C, et al. Detection rate of quadruple‐marker screening determined by clinical follow‐up and registry data in the statewide California program, July 2007 to February 2009. Prenatal Diagnosis 2011;31(9):901‐6. [PUBMED: 21706514] - PubMed
LabGenomics 2016
-
- LabGenomics Clinical Research Institute, Korea. www.momguard.com/eng/home.php?go=main (accessed 5 December 2016).
Langlois 2011
-
- Langlois S, Duncan A. Use of a DNA method, QF‐PCR, in the prenatal diagnosis of fetal aneuploidies. Journal of Obstetrics and Gynaecology Canada 2011;33(9):955‐60. [PUBMED: 21923994] - PubMed
Larion 2014
-
- Larion S, Warsof SL, Romary L, Mlynarczyk M, Peleg D, Abuhamad AZ. Association of combined first‐trimester screen and noninvasive prenatal testing on diagnostic procedures. Obstetrics and Gynecology 2014;123(6):1303‐10. [PUBMED: 24807333] - PubMed
Legare 2010
Legare 2011
-
- Legare F, St‐Jacques S, Gagnon S, Njoya M, Brisson M, Fremont P, et al. Prenatal screening for Down syndrome: a survey of willingness in women and family physicians to engage in shared decision‐making. Prenatal Diagnosis 2011;31(4):319‐26. [PUBMED: 21268046] - PubMed
Leggett 2010
LifeCodexx 2016
-
- LifeCodexx AG, Germany. lifecodexx.com/en/for‐physicians/methods‐technology/clinical‐validation/ (accessed 10 November 2016).
Lo 1997
-
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350(9076):485‐7. [PUBMED: 9274585] - PubMed
Lo 1999
Lun 2008
-
- Lun FM, Chiu RW, Allen Chan KC, Yeung Leung T, Kin Lau T, Dennis Lo YM. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clinical Chemistry 2008;54(10):1664‐72. [PUBMED: 18703764] - PubMed
Macaskill 2010
-
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: Analysing and presenting results. In: Deeks JJ, Bossuyt PM, Gatsonis C, (editors), Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0. The Cochrane Collaboration 2010. Available from: methods.cochrane.org/sdt/handbook‐dta‐reviews.
Mackie 2017
-
- Mackie FL, Hemming K, Allen S, Morris RK, Kilby MD. The accuracy of cell‐free fetal DNA‐based non‐invasive prenatal testing in singleton pregnancies: a systematic review and bivariate meta‐analysis. BJOG: an international journal of obstetrics and gynaecology 2017;124(1):32‐46. [PUBMED: 27245374] - PubMed
Mazzanti 1998
-
- Mazzanti L, Cacciari E. Congenital heart disease in patients with Turner's syndrome. Italian Study Group for Turner Syndrome (ISGTS). Journal of Pediatrics 1998;133(5):688‐92. [PUBMED: 9821430] - PubMed
MedlinePlus 2014
-
- Medical Encyclopedia, A service of the U.S. National Library of Medicine, National Institutes of Health. medlineplus.gov/mplusdictionary.html (updated daily, accessed 17 July 2014).
Mersy 2013
-
- Mersy E, Smits LJ, Winden LA, Die‐Smulders CE, Paulussen AD, Macville MV, et al. Noninvasive detection of fetal trisomy 21: systematic review and report of quality and outcomes of diagnostic accuracy studies performed between 1997 and 2012. Human Reproduction Update 2013;19(4):318‐29. [PUBMED: 23396607] - PubMed
Modi 2003
-
- Modi D, Berde P, Bhartiya D. Down syndrome: a study of chromosomal mosaicism. Reproductive Biomedicine Online 2003;6(4):499‐503. [PUBMED: 12831601] - PubMed
Natera 2016
-
- Natera, Inc, USA. www.natera.com/panorama‐test/clinical‐information (accessed 25 October 2016).
Newcombe 1998
-
- Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Statistics in Medicine 1998;17(8):873‐90. [PUBMED: 9595617] - PubMed
Nshimyumukiza 2017
-
- Nshimyumukiza L, Menon S, Hina H, Rousseau F, Reinharz D. Cell‐free DNA noninvasive prenatal screening for aneuploidy versus conventional screening: a systematic review of economic evaluations. Clinical Genetics in press. - PubMed
O'Leary 2013
-
- O'Leary P, Maxwell S, Murch A, Hendrie D. Prenatal screening for Down syndrome in Australia: costs and benefits of current and novel screening strategies. Australian & New Zealand Journal of Obstetrics & Gynaecology 2013;53(5):425‐33. [PUBMED: 24090461] - PubMed
Okun 2008
-
- Okun N, Summers AM, Hoffman B, Huang T, Winsor E, Chitayat D, et al. Prospective experience with integrated prenatal screening and first trimester combined screening for trisomy 21 in a large Canadian urban center. Prenatal Diagnosis 2008;28(11):987‐92. [PUBMED: 18925623] - PubMed
Papageorgiou 2012
Parker 2010
-
- Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004‐2006. Birth Defects Research. Part A, Clinical and Molecular Teratology 2010;88(12):1008‐16. [PUBMED: 20878909] - PubMed
Premaitha Health plc 2016
-
- Premaitha Health public limited company, UK. (accessed 15 November 2016).
Renshaw 2013
-
- Renshaw R, Ellis K, Jacobs P, Morris J. Antenatal screening for Down syndrome: a quantitative demonstration of the improvements over the past 20 years. Journal of Health Services Research & Policy 2013;18(4):195‐201. [PUBMED: 23864125] - PubMed
Rothberg 2011
-
- Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, et al. An integrated semiconductor device enabling non‐optical genome sequencing. Nature 2011;475(7356):348‐52. [PUBMED: 21776081] - PubMed
Rutter 2001
-
- Rutter CM, Gatsonis CA. A hierarchical regression approach to meta‐analysis of diagnostic test accuracy evaluations. Statistics in Medicine 2001;20(19):2865‐84. [PUBMED: 11568945] - PubMed
Saenger 1996
-
- Saenger P. Turner's syndrome. New England Journal of Medicine 1996;335(23):1749‐54. [PUBMED: 8929268] - PubMed
Samura 2003
-
- Samura O, Miharu N, Hyodo M, Honda H, Ohashi Y, Honda N, et al. Cell‐free fetal DNA in maternal circulation after amniocentesis. Clinical Chemistry 2003;49(7):1193‐5. [PUBMED: 12816922] - PubMed
Schoemaker 2008
-
- Schoemaker MJ, Swerdlow AJ, Higgins CD, Wright AF, Jacobs PA. Mortality in women with turner syndrome in Great Britain: a national cohort study. Journal of Clinical Endocrinology and Metabolism 2008;93(12):4735‐42. [PUBMED: 18812477] - PubMed
Sequenom 2016
-
- Sequenom, Inc. www.sequenom.com/tests/reproductive‐health/maternit21‐plus#provider‐refe... and www.sequenom.com/tests/reproductive‐health/visibilit#provider‐test‐details (accessed 25 October 2016).
Snijders 1999
-
- Snijders RJ, Sundberg K, Holzgreve W, Henry G, Nicolaides KH. Maternal age‐ and gestation‐specific risk for trisomy 21. Ultrasound in Obstetrics & Gynecology 1999;13(3):167‐70. [PUBMED: 10204206] - PubMed
South 2013
-
- South ST, Lee C, Lamb AN, Higgins AW, Kearney HM. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genetics in Medicine 2013;15(11):901‐9. [PUBMED: 24071793] - PubMed
St‐Jacques 2008
-
- St‐Jacques S, Grenier S, Charland M, Forest JC, Rousseau F, Legare F. Decisional needs assessment regarding Down syndrome prenatal testing: a systematic review of the perceptions of women, their partners and health professionals. Prenatal Diagnosis 2008;28(13):1183‐203. [PUBMED: 19097031] - PubMed
Stochholm 2006
-
- Stochholm K, Juul S, Juel K, Naeraa RW, Gravholt CH. Prevalence, incidence, diagnostic delay, and mortality in Turner syndrome. Journal of Clinical Endocrinology and Metabolism 2006;91(10):3897‐902. [PUBMED: 16849410] - PubMed
Stochholm 2010a
Stochholm 2010b
-
- Stochholm K, Juul S, Gravholt CH. Mortality and incidence in women with 47,XXX and variants. American Journal of Medical Genetics. Part a 2010;152A(2):367‐72. [PUBMED: 20101696] - PubMed
Summers 2007
-
- Summers AM, Langlois S, Wyatt P, Wilson RD, Society of Obstetricians and Gynaecologists of Canada. Prenatal screening for fetal aneuploidy. Journal of Obstetrics and Gynaecology Canada : JOGC 2007;29(2):146‐79. [PUBMED: 17346485] - PubMed
Sybert 2004
-
- Sybert VP, McCauley E. Turner's syndrome. New England Journal of Medicine 2004;351(12):1227‐38. [PUBMED: 15371580] - PubMed
Takwoingi 2015
Tartaglia 2010
Taylor‐Phillips 2016
Tiller 2014
-
- Tiller GE, Kershberg HB, Goff J, Coffeen C, Liao W, Sehnert AJ. Women's views and the impact of noninvasive prenatal testing on procedures in a managed care setting. Prenatal Diagnosis 2014;34:1‐6. [PUBMED: 25201151] - PubMed
Tyler 2004
-
- Tyler C, Edman JC. Down syndrome, Turner syndrome, and Klinefelter syndrome: primary care throughout the life span. Primary Care 2004;31(3):627‐48, x‐xi. [PUBMED: 15331252] - PubMed
Uematsu 2002
-
- Uematsu A, Yorifuji T, Muroi J, Kawai M, Mamada M, Kaji M, et al. Parental origin of normal X chromosomes in Turner syndrome patients with various karyotypes: implications for the mechanism leading to generation of a 45,X karyotype. American Journal of Medical Genetics 2002;111(2):134‐9. [PUBMED: 12210339] - PubMed
UK Screening Glossary 2012
-
- United Kingdom National Screening Committee Glossary. webarchive.nationalarchives.gov.uk/20150408175925/http://www.screening.n... (Last reviewed June 2012, accessed 18 November 2013).
Van Riper 2001
-
- Riper M, Cohen WI. Caring for children with Down syndrome and their families. Journal of Pediatric Health Care 2001;15(3):123‐31. [PUBMED: 11353361] - PubMed
Vora 2010
Wald 2005
-
- Wald NJ, Rodeck C, Hackshaw AK, Rudnicka A. SURUSS in perspective. Seminars in Perinatology 2005;29(4):225‐35. [PUBMED: 16104673] - PubMed
Weijerman 2010
Wellesley 2012
Whiting 2011
-
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529‐36. [PUBMED: 22007046] - PubMed
Wilson 2007
-
- Wilson RD, Langlois S, Johnson JA. Mid‐trimester amniocentesis fetal loss rate. Journal of Obstetrics and Gynaecology Canada : JOGC 2007;29(7):586‐95. [PUBMED: 17623573] - PubMed
Wright 2009
-
- Wright C. Cell‐free fetal nucleic acids for non‐invasive prenatal diagnosis, Report of the UK expert working group. PHG foundation (2009).
Wu 2013a
-
- Wu J, Springett A, Morris JK. Survival of trisomy 18 (Edwards syndrome) and trisomy 13 (Patau Syndrome) in England and Wales: 2004‐2011. American Journal of Medical Genetics. Part a 2013;161(10):2512‐8. [PUBMED: 23949924] - PubMed
Wu 2013b
Yaron 2016
-
- Yaron Y. The implications of non‐invasive prenatal testing failures: a review of an under‐discussed phenomenon. Prenatal Diagnosis 2016;36(5):391‐6. [PUBMED: 26941176] - PubMed
Yu 2013
-
- Yu SC, Lee SW, Jiang P, Leung TY, Chan KC, Chiu RW, et al. High‐resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clinical Chemistry 2013;59(8):1228‐37. [PUBMED: 23603797] - PubMed
References to other published versions of this review
Badeau 2015
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

