New aspects of RNA-based regulation by Hfq and its partner sRNAs
- PMID: 29125938
- PMCID: PMC10367044
- DOI: 10.1016/j.mib.2017.10.014
New aspects of RNA-based regulation by Hfq and its partner sRNAs
Abstract
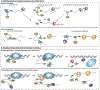
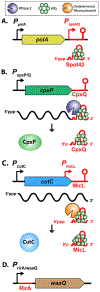
Hfq, an RNA chaperone, promotes the pairing of small RNAs (sRNAs) to target mRNAs, mediating post-transcriptional regulation of mRNA stability and translation. This regulation contributes to bacterial adaptation during stress and pathogenesis. Recent advances in sequencing techniques demonstrate the presence of sRNAs encoded not only in intergenic regions but also from the 3' and 5' UTRs of mRNAs, expanding sRNA regulatory networks. Additional layers of regulation by Hfq and its associated RNAs continue to be found. Newly identified RNA sponges modulate the activity of some sRNAs. A subset of sRNAs are proving to be bifunctional, able to pair with targets and also encoding small ORFs or binding other RNA binding proteins, such as CsrA. In addition, there are accumulating examples of Hfq inhibiting mRNA translation in the absence of sRNAs.
Published by Elsevier Ltd.
Figures
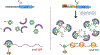
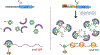
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

