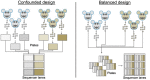
Experimental design for single-cell RNA sequencing
- PMID: 29126257
- PMCID: PMC6063265
- DOI: 10.1093/bfgp/elx035
Experimental design for single-cell RNA sequencing
Abstract
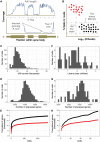
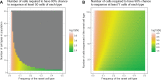
Single-cell RNA sequencing (scRNA-seq) has opened new avenues for the characterization of heterogeneity in a large variety of cellular systems. As this is a relatively new technique, the field is fast evolving. Here, we discuss general considerations in experimental design and the two most popular approaches, plate-based Smart-Seq2 and microdroplet-based scRNA-seq at the example of 10x Chromium. We discuss advantages and disadvantages of both methods and point out major factors to consider in designing successful experiments.
Figures
References
-
- Picelli S, Faridani OR, Björklund ÅK, et al.Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014;9(1):171–81. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

