RNA-sequencing analysis of lung primary fibroblast response to eosinophil-degranulation products predicts downstream effects on inflammation, tissue remodeling and lipid metabolism
- PMID: 29126429
- PMCID: PMC5681771
- DOI: 10.1186/s12931-017-0669-8
RNA-sequencing analysis of lung primary fibroblast response to eosinophil-degranulation products predicts downstream effects on inflammation, tissue remodeling and lipid metabolism
Abstract
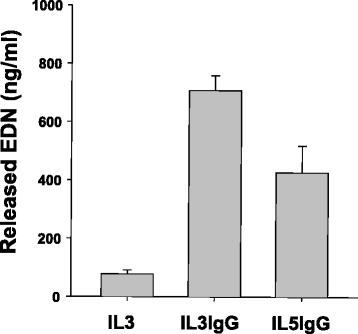
Background: The association of eosinophils with inflammation and tissue remodeling is at least partially due to their release of toxic granule proteins and other mediators, including cytokines. Tissue remodeling and consequent functional defects are affected by activity of connective tissue fibroblasts. Exaggerated fibroblast activation, accumulation and change of phenotype may lead to fibrosis and loss of tissue function. So far, little information has been reported on how eosinophils affect inflammation and tissue remodeling via the activation of fibroblasts. We have recently shown that eosinophil activation with IL-3 led to a robust eosinophil degranulation on immunoglobin-G (IgG) coated plates. Thus, in the present study, we analyze the effects of IL-3-activated eosinophil degranulation products on primary human lung fibroblasts (HLF) using whole transcriptome sequencing.
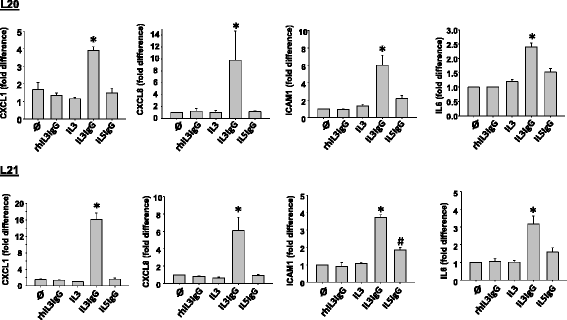
Methods: Conditioned media was obtained from eosinophils that were pre-activated with IL-3 or IL-5 and subsequently cultured for 6 h on IgG to induce degranulation. This conditioned media was added on human lung fibroblasts (HLF) for 24 h and the cell lysates were then subjected to whole transcriptome sequencing to identify global changes in gene expression. Differentially expressed genes were analyzed using the Ingenuity Pathway Analysis (IPA), and validated by qPCR.
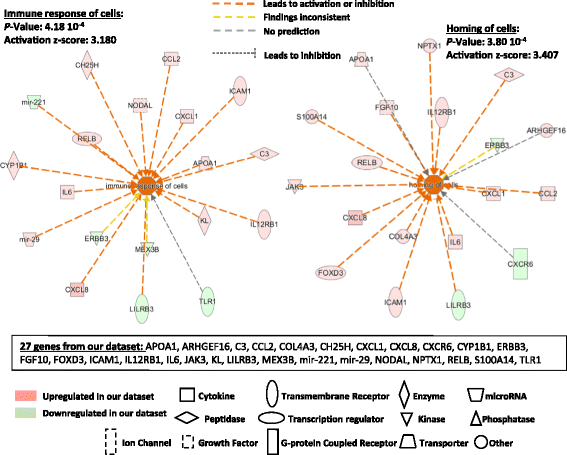
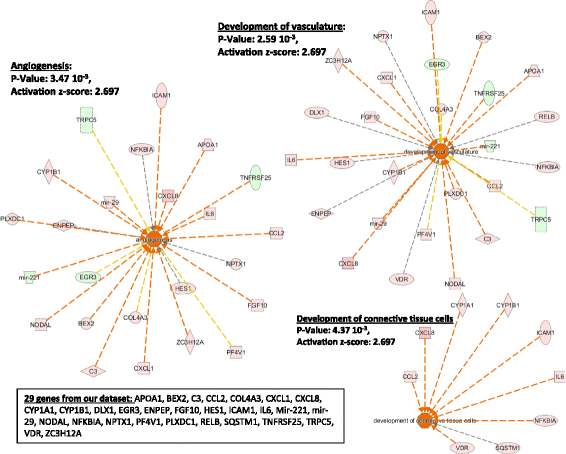
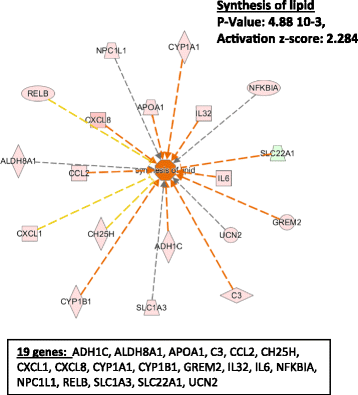
Results: In HLF, the expression level of 300 genes was changed by conditioned media from IL-3-activated eosinophils compared to control fibroblast cultures. Among these 300 genes, the expression level of 35 genes coding for known proteins was upregulated by IL-3- versus IL-5-pre-activated eosinophils. Of the 35 upregulated genes, IPA identified C3, CH25H, CXCL1, CXCL8, CYP1A1, ICAM1, IL6 and UCN2 as having downstream functions on inflammation, tissue remodeling and lipid synthesis. This analysis combined with previous RNA sequencing analyses of eosinophils suggest IL-1ß, OSM and TNFSF12 as potential upstream regulators of fibroblasts.
Conclusions: This study has identified several novel pro-inflammatory and pro-remodeling mediators produced by fibroblasts in response to activated eosinophils. These findings may have significant implications on the role of eosinophil/fibroblast interactions in eosinophilic disorders.
Keywords: Degranulation; Eosinophil; Fibroblast; IgG; Il-3; Inflammation; Lipid metabolism; Lung; RNA-sequencing; Tissue remodeling.
Conflict of interest statement
Ethics approval and consent to participate
The study protocol was approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board (IRB). The use of blood eosinophils was approved by the IRB committee’s reference number: 2014–1481. The use of pulmonary fibroblasts was approved by IRB committee’s number: 2012–0061.
Consent for publication
Not applicable
Competing interests
Dr. Nizar Jarjour is a consultant for Astra Zeneca in the areas of developing new therapeutic for asthma and COPD. The other authors declare that they have no competing interests related to this work.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Malinovschi A, Fonseca JA, Jacinto T, Alving K, Janson C. Exhaled nitric oxide levels and blood eosinophil counts independently associate with wheeze and asthma events in National Health and nutrition examination survey subjects. J Allergy Clin Immunol. 2013;132:821–827. doi: 10.1016/j.jaci.2013.06.007. - DOI - PubMed
-
- Bel EH, Ortega HG, Pavord ID: Glucocorticoids and mepolizumab in eosinophilic asthma REPLY. N Engl J Med 2014, 371:2434-2434. - PubMed
MeSH terms
Substances
Grants and funding
- P30 CA014520/Comprehensive Cancer Center, University of Wisconsin
- UL1TR000427/National Center for Advancing Translational Sciences
- NIH HL088594/National Heart, Lung, and Blood Institute
- UL1 TR000427/TR/NCATS NIH HHS/United States
- P30 CA014520/CA/NCI NIH HHS/United States
- K08 HL093367/HL/NHLBI NIH HHS/United States
- P01 HL088594/HL/NHLBI NIH HHS/United States
- Pulmonary Fibrosis Foundation Research Grant/Coalition for Pulmonary Fibrosis
- ATS Foundation/American Thoracic Society
- Development Funding/University of Wisconsin-Madison
- R03 HL136795/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

