HIV-1 NC-induced stress granule assembly and translation arrest are inhibited by the dsRNA binding protein Staufen1
- PMID: 29127210
- PMCID: PMC5769749
- DOI: 10.1261/rna.064618.117
HIV-1 NC-induced stress granule assembly and translation arrest are inhibited by the dsRNA binding protein Staufen1
Abstract
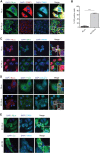
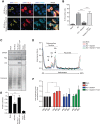
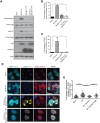
The nucleocapsid (NC) is an N-terminal protein derived from the HIV-1 Gag precursor polyprotein, pr55Gag NC possesses key functions at several pivotal stages of viral replication. For example, an interaction between NC and the host double-stranded RNA-binding protein Staufen1 was shown to regulate several steps in the viral replication cycle, such as Gag multimerization and genomic RNA encapsidation. In this work, we observed that the overexpression of NC leads to the induction of stress granule (SG) assembly. NC-mediated SG assembly was unique as it was resistant to the SG blockade imposed by the HIV-1 capsid (CA), as shown in earlier work. NC also reduced host cell mRNA translation, as judged by a puromycylation assay of de novo synthesized proteins, and this was recapitulated in polysome profile analyses. Virus production was also found to be significantly reduced. Finally, Staufen1 expression completely rescued the blockade to NC-mediated SG assembly, global mRNA translation as well as virus production. NC expression also resulted in the phosphorylation of protein kinase R (PKR) and eIF2α, and this was inhibited with Staufen1 coexpression. This work sheds light on an unexpected function of NC in host cell translation. A comprehensive understanding of the molecular mechanisms by which a fine balance of the HIV-1 structural proteins NC and CA act in concert with host proteins such as Staufen1 to modulate the host stress response will aid in the development of new antiviral therapeutics.
Keywords: HIV-1; Staufen1; mRNA translation; nucleocapsid; stress granules.
© 2018 Rao et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





Similar articles
-
Newcastle disease virus induces stable formation of bona fide stress granules to facilitate viral replication through manipulating host protein translation.FASEB J. 2017 Apr;31(4):1337-1353. doi: 10.1096/fj.201600980R. Epub 2016 Dec 23. FASEB J. 2017. PMID: 28011649
-
Mouse Norovirus Infection Arrests Host Cell Translation Uncoupled from the Stress Granule-PKR-eIF2α Axis.mBio. 2019 Jun 18;10(3):e00960-19. doi: 10.1128/mBio.00960-19. mBio. 2019. PMID: 31213553 Free PMC article.
-
Zika Virus Subverts Stress Granules To Promote and Restrict Viral Gene Expression.J Virol. 2019 May 29;93(12):e00520-19. doi: 10.1128/JVI.00520-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30944179 Free PMC article.
-
Rasputin a decade on and more promiscuous than ever? A review of G3BPs.Biochim Biophys Acta Mol Cell Res. 2019 Mar;1866(3):360-370. doi: 10.1016/j.bbamcr.2018.09.001. Epub 2018 Sep 5. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30595162 Free PMC article. Review.
-
Cataloguing and Selection of mRNAs Localized to Dendrites in Neurons and Regulated by RNA-Binding Proteins in RNA Granules.Biomolecules. 2020 Jan 22;10(2):167. doi: 10.3390/biom10020167. Biomolecules. 2020. PMID: 31978946 Free PMC article. Review.
Cited by
-
Human immunodeficiency virus-1 core: The Trojan horse in virus-host interaction.Front Microbiol. 2022 Aug 29;13:1002476. doi: 10.3389/fmicb.2022.1002476. eCollection 2022. Front Microbiol. 2022. PMID: 36106078 Free PMC article. Review.
-
Strategies for Success. Viral Infections and Membraneless Organelles.Front Cell Infect Microbiol. 2019 Oct 11;9:336. doi: 10.3389/fcimb.2019.00336. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31681621 Free PMC article. Review.
-
Pan-retroviral Nucleocapsid-Mediated Phase Separation Regulates Genomic RNA Positioning and Trafficking.Cell Rep. 2020 Apr 21;31(3):107520. doi: 10.1016/j.celrep.2020.03.084. Cell Rep. 2020. PMID: 32320662 Free PMC article.
-
A little less aggregation a little more replication: Viral manipulation of stress granules.Wiley Interdiscip Rev RNA. 2023 Jan;14(1):e1741. doi: 10.1002/wrna.1741. Epub 2022 Jun 16. Wiley Interdiscip Rev RNA. 2023. PMID: 35709333 Free PMC article. Review.
-
HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles.RNA. 2019 Jun;25(6):727-736. doi: 10.1261/rna.069351.118. Epub 2019 Mar 22. RNA. 2019. PMID: 30902835 Free PMC article.
References
-
- Abrahamyan L, Chatel-Chaix L, Ajamian L, Milev M, Monette A, Clément JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, et al. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci 123: 369–383. - PubMed
-
- Anderson P, Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
