Whole Exome Sequencing of Patients with Steroid-Resistant Nephrotic Syndrome
- PMID: 29127259
- PMCID: PMC5753307
- DOI: 10.2215/CJN.04120417
Whole Exome Sequencing of Patients with Steroid-Resistant Nephrotic Syndrome
Abstract
Background and objectives: Steroid-resistant nephrotic syndrome overwhelmingly progresses to ESRD. More than 30 monogenic genes have been identified to cause steroid-resistant nephrotic syndrome. We previously detected causative mutations using targeted panel sequencing in 30% of patients with steroid-resistant nephrotic syndrome. Panel sequencing has a number of limitations when compared with whole exome sequencing. We employed whole exome sequencing to detect monogenic causes of steroid-resistant nephrotic syndrome in an international cohort of 300 families.
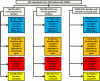
Design, setting, participants, & measurements: Three hundred thirty-five individuals with steroid-resistant nephrotic syndrome from 300 families were recruited from April of 1998 to June of 2016. Age of onset was restricted to <25 years of age. Exome data were evaluated for 33 known monogenic steroid-resistant nephrotic syndrome genes.
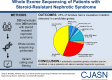
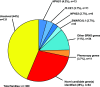
Results: In 74 of 300 families (25%), we identified a causative mutation in one of 20 genes known to cause steroid-resistant nephrotic syndrome. In 11 families (3.7%), we detected a mutation in a gene that causes a phenocopy of steroid-resistant nephrotic syndrome. This is consistent with our previously published identification of mutations using a panel approach. We detected a causative mutation in a known steroid-resistant nephrotic syndrome gene in 38% of consanguineous families and in 13% of nonconsanguineous families, and 48% of children with congenital nephrotic syndrome. A total of 68 different mutations were detected in 20 of 33 steroid-resistant nephrotic syndrome genes. Fifteen of these mutations were novel. NPHS1, PLCE1, NPHS2, and SMARCAL1 were the most common genes in which we detected a mutation. In another 28% of families, we detected mutations in one or more candidate genes for steroid-resistant nephrotic syndrome.
Conclusions: Whole exome sequencing is a sensitive approach toward diagnosis of monogenic causes of steroid-resistant nephrotic syndrome. A molecular genetic diagnosis of steroid-resistant nephrotic syndrome may have important consequences for the management of treatment and kidney transplantation in steroid-resistant nephrotic syndrome.
Keywords: Child; Exome; Humans; Kidney Failure, Chronic; Mutation; Nephrosis, congenital; Phenotype; Renal Insufficiency, Chronic; genetic renal disease; kidney transplantation; molecular genetics; nephrotic syndrome; pediatric.
Copyright © 2018 by the American Society of Nephrology.
Figures


References
-
- Primary nephrotic syndrome in children: Clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A Report of the International Study of Kidney Disease in Children. Kidney Int 20: 765–771, 1981 - PubMed
-
- Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA: Contributions of the transplant registry: The 2006 annual report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr Transplant 11: 366–373, 2007 - PubMed
-
- Cheong HI, Han HW, Park HW, Ha IS, Han KS, Lee HS, Kim SJ, Choi Y: Early recurrent nephrotic syndrome after renal transplantation in children with focal segmental glomerulosclerosis. Nephrol Dial Transplant 15: 78–81, 2000 - PubMed
-
- Senggutuvan P, Cameron JS, Hartley RB, Rigden S, Chantler C, Haycock G, Williams DG, Ogg C, Koffman G: Recurrence of focal segmental glomerulosclerosis in transplanted kidneys: Analysis of incidence and risk factors in 59 allografts. Pediatr Nephrol 4: 21–28, 1990 - PubMed
-
- Hamiwka LA, Midgley JP, Wade AW, Martz KL, Grisaru S. Outcomes of kidney transplantation in children with nephronophthisis: an analysis of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Registry. Pediatr Transplant 12: 878–882, 2008 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

