Sequencing and characterization of the complete mitochondrial genome of Japanese Swellshark (Cephalloscyllium umbratile)
- PMID: 29127415
- PMCID: PMC5681689
- DOI: 10.1038/s41598-017-15702-0
Sequencing and characterization of the complete mitochondrial genome of Japanese Swellshark (Cephalloscyllium umbratile)
Abstract
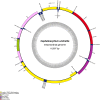
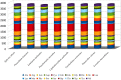
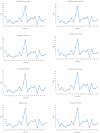
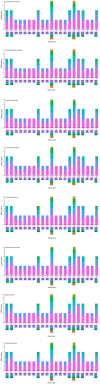
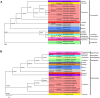
To further comprehend the genome features of Cephalloscyllium umbratile (Carcharhiniformes), an endangered species, the complete mitochondrial DNA (mtDNA) was firstly sequenced and annotated. The full-length mtDNA of C. umbratile was 16,697 bp and contained ribosomal RNA (rRNA) genes, 13 protein-coding genes (PCGs), 23 transfer RNA (tRNA) genes, and a major non-coding control region. Each PCG was initiated by an authoritative ATN codon, except for COX1 initiated by a GTG codon. Seven of 13 PCGs had a typical TAA termination codon, while others terminated with a single T or TA. Moreover, the relative synonymous codon usage of the 13 PCGs was consistent with that of other published Carcharhiniformes. All tRNA genes had typical clover-leaf secondary structures, except for tRNA-Ser (GCT), which lacked the dihydrouridine 'DHU' arm. Furthermore, the analysis of the average Ka/Ks in the 13 PCGs of three Carcharhiniformes species indicated a strong purifying selection within this group. In addition, phylogenetic analysis revealed that C. umbratile was closely related to Glyphis glyphis and Glyphis garricki. Our data supply a useful resource for further studies on genetic diversity and population structure of C. umbratile.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Complete mitochondrial genome of the Critically Endangered speartooth shark Glyphis glyphis (Carcharhiniformes: Carcharhinidae).Mitochondrial DNA. 2014 Dec;25(6):431-2. doi: 10.3109/19401736.2013.809443. Epub 2013 Jul 16. Mitochondrial DNA. 2014. PMID: 23859050
-
Complete mitogenomic sequence of the Critically Endangered Northern River Shark Glyphis garricki (Carcharhiniformes: Carcharhinidae).Mitochondrial DNA. 2015;26(6):855-6. doi: 10.3109/19401736.2013.861428. Epub 2014 Jan 10. Mitochondrial DNA. 2015. PMID: 24409900
-
Molecular characterization and phylogenetic analyses of the mitogenome of Wan-Xi white goose, a native goose breed in China.BMC Genom Data. 2025 May 13;26(1):34. doi: 10.1186/s12863-025-01326-1. BMC Genom Data. 2025. PMID: 40360978 Free PMC article.
-
The Complete Mitochondrial Genome of the Freshwater Fish Onychostoma ovale (Cypriniformes, Cyprinidae): Genome Characterization and Phylogenetic Analysis.Genes (Basel). 2023 Jun 6;14(6):1227. doi: 10.3390/genes14061227. Genes (Basel). 2023. PMID: 37372406 Free PMC article.
-
Complete mitochondrial genome and the phylogenetic position of the Blotchy swell shark Cephaloscyllium umbratile.Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Jul;27(4):3045-7. doi: 10.3109/19401736.2015.1063127. Epub 2015 Aug 10. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26258506
Cited by
-
Mitogenomic Architecture of Atlantic Emperor Lethrinus atlanticus (Actinopterygii: Spariformes): Insights into the Lineage Diversification in Atlantic Ocean.Int J Mol Sci. 2024 Oct 4;25(19):10700. doi: 10.3390/ijms251910700. Int J Mol Sci. 2024. PMID: 39409028 Free PMC article.
-
Detailed characterization of the complete mitochondrial genome of the oceanic whitetip shark Carcharhinus longimanus (Poey, 1861).Mol Biol Rep. 2024 Jul 19;51(1):826. doi: 10.1007/s11033-024-09780-3. Mol Biol Rep. 2024. PMID: 39030452 Free PMC article.
-
Characterization of 11 complete mitochondrial genomes in Nudibranchia (Mollusca, Gastropoda).Zookeys. 2025 Jul 4;1244:61-86. doi: 10.3897/zookeys.1244.139617. eCollection 2025. Zookeys. 2025. PMID: 40654599 Free PMC article.
-
Comparative mitogenomics and phylogenetics of the family Carangidae with special emphasis on the mitogenome of the Indian Scad Decapterus russelli.Sci Rep. 2022 Apr 4;12(1):5642. doi: 10.1038/s41598-022-09636-5. Sci Rep. 2022. PMID: 35379869 Free PMC article.
-
Comparative Analysis of Complete Mitochondrial Genomes of Five Chromodorididae Species (Nudibranchia:Doridina).Biochem Genet. 2025 Aug;63(4):3347-3362. doi: 10.1007/s10528-024-10878-3. Epub 2024 Jul 2. Biochem Genet. 2025. PMID: 38954214
References
-
- Iglésias, S., Tanaka, S. & Nakaya, K. Cephaloscyllium umbratile. The IUCN Red List of Threatened Species: e.T161724A5488954. 10.2305/IUCN.UK.2009-2.RLTS.T161724A5488954.en (2009).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

