Epigenetics in diabetic nephropathy, immunity and metabolism
- PMID: 29128937
- PMCID: PMC6448927
- DOI: 10.1007/s00125-017-4490-1
Epigenetics in diabetic nephropathy, immunity and metabolism
Abstract
When it comes to the epigenome, there is a fine line between clarity and confusion-walk that line and you will discover another fascinating level of transcription control. With the genetic code representing the cornerstone of rules for information that is encoded to proteins somewhere above the genome level there is a set of rules by which chemical information is also read. These epigenetic modifications show a different side of the genetic code that is diverse and regulated, hence modifying genetic transcription transiently, ranging from short- to long-term alterations. While this complexity brings exquisite control it also poses a formidable challenge to efforts to decode mechanisms underlying complex disease. Recent technological and computational advances have improved unbiased acquisition of epigenomic patterns to improve our understanding of the complex chromatin landscape. Key to resolving distinct chromatin signatures of diabetic complications is the identification of the true physiological targets of regulatory proteins, such as reader proteins that recognise, writer proteins that deposit and eraser proteins that remove specific chemical moieties. But how might a diverse group of proteins regulate the diabetic landscape from an epigenomic perspective? Drawing from an ever-expanding compendium of experimental and clinical studies, this review details the current state-of-play and provides a perspective of chromatin-dependent mechanisms implicated in diabetic complications, with a special focus on diabetic nephropathy. We hypothesise a codified signature of the diabetic epigenome and provide examples of prime candidates for chemical modification. As for the pharmacological control of epigenetic marks, we explore future strategies to expedite and refine the search for clinically relevant discoveries. We also consider the challenges associated with therapeutic strategies targeting epigenetic pathways.
Keywords: Chromatin; Diabetes; Diabetic complications; Diabetic nephropathy; EWAS; Epigenetics; Histone; Innate immune memory; Vascular.
Conflict of interest statement
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
All authors were responsible for drafting the article and revising it critically for important intellectual content. All authors approved the version to be published.
Figures
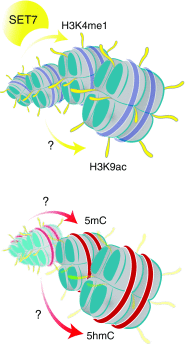
References
-
- DCCT/EDIC The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. N Engl J Med. 2000;342:381–389. doi: 10.1056/NEJM200002103420603. - DOI - PMC - PubMed
-
- Hammes HP, Klinzing I, Wiegand S, Bretzel RG, Cohen AM, Federlin K. Islet transplantation inhibits diabetic retinopathy in the sucrose-fed diabetic Cohen rat. Invest Ophthalmol Vis Sci. 1993;34:2092–2096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

