Functional Consequences of CHRNA7 Copy-Number Alterations in Induced Pluripotent Stem Cells and Neural Progenitor Cells
- PMID: 29129316
- PMCID: PMC5812918
- DOI: 10.1016/j.ajhg.2017.09.024
Functional Consequences of CHRNA7 Copy-Number Alterations in Induced Pluripotent Stem Cells and Neural Progenitor Cells
Abstract
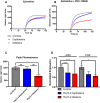
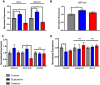
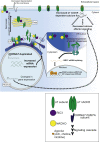
Copy-number variants (CNVs) of chromosome 15q13.3 manifest clinically as neuropsychiatric disorders with variable expressivity. CHRNA7, encoding for the α7 nicotinic acetylcholine receptor (nAChR), has been suggested as a candidate gene for the phenotypes observed. Here, we used induced pluripotent stem cells (iPSCs) and neural progenitor cells (NPCs) derived from individuals with heterozygous 15q13.3 deletions and heterozygous 15q13.3 duplications to investigate the CHRNA7-dependent molecular consequences of the respective CNVs. Unexpectedly, both deletions and duplications lead to decreased α7 nAChR-associated calcium flux. For deletions, this decrease in α7 nAChR-dependent calcium flux is expected due to haploinsufficiency of CHRNA7. For duplications, we found that increased expression of CHRNA7 mRNA is associated with higher expression of nAChR-specific and resident ER chaperones, indicating increased ER stress. This is likely a consequence of inefficient chaperoning and accumulation of α7 subunits in the ER, as opposed to being incorporated into functional α7 nAChRs at the cell membrane. Here, we showed that α7 nAChR-dependent calcium signal cascades are downregulated in both 15q13.3 deletion and duplication NPCs. While it may seem surprising that genomic changes in opposite direction have consequences on downstream pathways that are in similar direction, it aligns with clinical data, which suggest that both individuals with deletions and duplications of 15q13.3 manifest neuropsychiatric disease and cognitive deficits.
Keywords: 15q13.3 CNVs; CHRNA7; NPCs; iPSCs; neurodevelopmental disorders; neuropsychiatric disorders.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Spielmann M., Reichelt G., Hertzberg C., Trimborn M., Mundlos S., Horn D., Klopocki E. Homozygous deletion of chromosome 15q13.3 including CHRNA7 causes severe mental retardation, seizures, muscular hypotonia, and the loss of KLF13 and TRPM1 potentially cause macrocytosis and congenital retinal dysfunction in siblings. Eur. J. Med. Genet. 2011;54:e441–e445. - PubMed
-
- Lepichon J.B., Bittel D.C., Graf W.D., Yu S. A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7, and other homozygously deleted genes. Am. J. Med. Genet. A. 2010;152A:1300–1304. - PubMed
-
- Masurel-Paulet A., Drumare I., Holder M., Cuisset J.M., Vallée L., Defoort S., Bourgois B., Pernes P., Cuvellier J.C., Huet F. Further delineation of eye manifestations in homozygous 15q13.3 microdeletions including TRPM1: a differential diagnosis of ceroid lipofuscinosis. Am. J. Med. Genet. A. 2014;164A:1537–1544. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

