Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead
- PMID: 29129718
- PMCID: PMC5777153
- DOI: 10.1016/j.chembiol.2017.09.010
Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead
Abstract
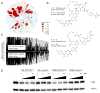
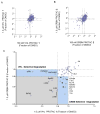
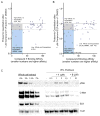
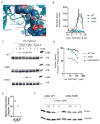
Inhibiting protein function selectively is a major goal of modern drug discovery. Here, we report a previously understudied benefit of small molecule proteolysis-targeting chimeras (PROTACs) that recruit E3 ubiquitin ligases to target proteins for their ubiquitination and subsequent proteasome-mediated degradation. Using promiscuous CRBN- and VHL-recruiting PROTACs that bind >50 kinases, we show that only a subset of bound targets is degraded. The basis of this selectivity relies on protein-protein interactions between the E3 ubiquitin ligase and the target protein, as illustrated by engaged proteins that are not degraded as a result of unstable ternary complexes with PROTAC-recruited E3 ligases. In contrast, weak PROTAC:target protein affinity can be stabilized by high-affinity target:PROTAC:ligase trimer interactions, leading to efficient degradation. This study highlights design guidelines for generating potent PROTACs as well as possibilities for degrading undruggable proteins immune to traditional small-molecule inhibitors.
Keywords: CRBN; MAPK/p38; PROTACs; VHL; chemical biology; protein degradation; protein-protein interactions; proteomics; selective degradation; ternary complex.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
Comment in
-
Try Me: Promiscuous Inhibitors Still Allow for Selective Targeted Protein Degradation.Cell Chem Biol. 2018 Jan 18;25(1):4-6. doi: 10.1016/j.chembiol.2018.01.004. Cell Chem Biol. 2018. PMID: 29351837
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

