Microbial Ecology along the Gastrointestinal Tract
- PMID: 29129876
- PMCID: PMC5745014
- DOI: 10.1264/jsme2.ME17017
Microbial Ecology along the Gastrointestinal Tract
Abstract
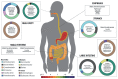
The ecosystem of the human gastrointestinal (GI) tract traverses a number of environmental, chemical, and physical conditions because it runs from the oral cavity to the anus. These differences in conditions along with food or other ingested substrates affect the composition and density of the microbiota as well as their functional roles by selecting those that are the most suitable for that environment. Previous studies have mostly focused on Bacteria, with the number of studies conducted on Archaea, Eukarya, and Viruses being limited despite their important roles in this ecosystem. Furthermore, due to the challenges associated with collecting samples directly from the inside of humans, many studies are still exploratory, with a primary focus on the composition of microbiomes. Thus, mechanistic studies to investigate functions are conducted using animal models. However, differences in physiology and microbiomes need to be clarified in order to aid in the translation of animal model findings into the context of humans. This review will highlight Bacteria, Archaea, Fungi, and Viruses, discuss differences along the GI tract of healthy humans, and perform comparisons with three common animal models: rats, mice, and pigs.
Keywords: Microbiome; animal models; diet; human gastrointestinal (GI) tract; mycobiome; virome.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

