A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells
- PMID: 29129916
- PMCID: PMC5682285
- DOI: 10.1038/s41467-017-01744-5
A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells
Abstract
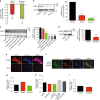
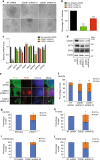
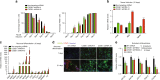
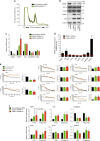
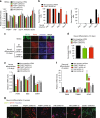
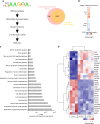
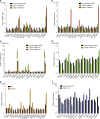
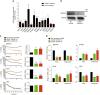
While the transcriptional network of human embryonic stem cells (hESCs) has been extensively studied, relatively little is known about how post-transcriptional modulations determine hESC function. RNA-binding proteins play central roles in RNA regulation, including translation and turnover. Here we show that the RNA-binding protein CSDE1 (cold shock domain containing E1) is highly expressed in hESCs to maintain their undifferentiated state and prevent default neural fate. Notably, loss of CSDE1 accelerates neural differentiation and potentiates neurogenesis. Conversely, ectopic expression of CSDE1 impairs neural differentiation. We find that CSDE1 post-transcriptionally modulates core components of multiple regulatory nodes of hESC identity, neuroectoderm commitment and neurogenesis. Among these key pro-neural/neuronal factors, CSDE1 binds fatty acid binding protein 7 (FABP7) and vimentin (VIM) mRNAs, as well as transcripts involved in neuron projection development regulating their stability and translation. Thus, our results uncover CSDE1 as a central post-transcriptional regulator of hESC identity and neurogenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Csde1 binds transcripts involved in protein homeostasis and controls their expression in an erythroid cell line.Sci Rep. 2018 Feb 8;8(1):2628. doi: 10.1038/s41598-018-20518-7. Sci Rep. 2018. PMID: 29422612 Free PMC article.
-
Strap associates with Csde1 and affects expression of select Csde1-bound transcripts.PLoS One. 2018 Aug 23;13(8):e0201690. doi: 10.1371/journal.pone.0201690. eCollection 2018. PLoS One. 2018. PMID: 30138317 Free PMC article.
-
Identification of TIA1 mRNA targets during human neuronal development.Mol Biol Rep. 2021 Sep;48(9):6349-6361. doi: 10.1007/s11033-021-06634-0. Epub 2021 Aug 19. Mol Biol Rep. 2021. PMID: 34410578 Free PMC article.
-
The role of CSDE1 in translational reprogramming and human diseases.Cell Commun Signal. 2020 Jan 27;18(1):14. doi: 10.1186/s12964-019-0496-2. Cell Commun Signal. 2020. PMID: 31987048 Free PMC article. Review.
-
Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons.Neuroscience. 2013 Sep 17;248:499-528. doi: 10.1016/j.neuroscience.2013.05.042. Epub 2013 May 30. Neuroscience. 2013. PMID: 23727006 Free PMC article. Review.
Cited by
-
CSDE1 controls gene expression through the miRNA-mediated decay machinery.Life Sci Alliance. 2020 Mar 11;3(4):e201900632. doi: 10.26508/lsa.201900632. Print 2020 Apr. Life Sci Alliance. 2020. PMID: 32161113 Free PMC article.
-
Maternal Protein Restriction in Rats Alters the Expression of Genes Involved in Mitochondrial Metabolism and Epitranscriptomics in Fetal Hypothalamus.Nutrients. 2020 May 19;12(5):1464. doi: 10.3390/nu12051464. Nutrients. 2020. PMID: 32438566 Free PMC article.
-
The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington's disease patients.Nat Commun. 2018 Jul 23;9(1):2886. doi: 10.1038/s41467-018-05320-3. Nat Commun. 2018. PMID: 30038412 Free PMC article.
-
Emerging Roles of RNA-Binding Proteins in Neurodevelopment.J Dev Biol. 2022 Jun 10;10(2):23. doi: 10.3390/jdb10020023. J Dev Biol. 2022. PMID: 35735914 Free PMC article. Review.
-
Challenges in Paragangliomas and Pheochromocytomas: from Histology to Molecular Immunohistochemistry.Endocr Pathol. 2021 Jun;32(2):228-244. doi: 10.1007/s12022-021-09675-0. Epub 2021 Mar 25. Endocr Pathol. 2021. PMID: 33768452 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

