Towards a genomics-informed, real-time, global pathogen surveillance system
- PMID: 29129921
- PMCID: PMC7097748
- DOI: 10.1038/nrg.2017.88
Towards a genomics-informed, real-time, global pathogen surveillance system
Abstract
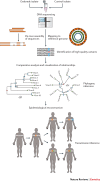
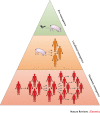
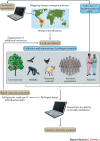
The recent Ebola and Zika epidemics demonstrate the need for the continuous surveillance, rapid diagnosis and real-time tracking of emerging infectious diseases. Fast, affordable sequencing of pathogen genomes - now a staple of the public health microbiology laboratory in well-resourced settings - can affect each of these areas. Coupling genomic diagnostics and epidemiology to innovative digital disease detection platforms raises the possibility of an open, global, digital pathogen surveillance system. When informed by a One Health approach, in which human, animal and environmental health are considered together, such a genomics-based system has profound potential to improve public health in settings lacking robust laboratory capacity.
Conflict of interest statement
J.L.G. declares no competing interests. N.J.L. has received travel expenses and accommodation and an honorarium payment from Oxford Nanopore Technologies to speak at organized symposia. N.J.L. is a member of the Oxford Nanopore MinION Access Programme and has received reagents for nanopore sequencing free of charge.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

