The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding
- PMID: 29129932
- PMCID: PMC5682278
- DOI: 10.1038/s41467-017-01542-z
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding
Abstract
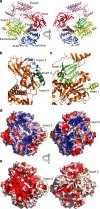
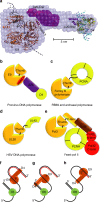
Vaccinia virus (VACV), the prototype member of the Poxviridae, replicates in the cytoplasm of an infected cell. The catalytic subunit of the DNA polymerase E9 binds the heterodimeric processivity factor A20/D4 to form the functional polymerase holoenzyme. Here we present the crystal structure of full-length E9 at 2.7 Å resolution that permits identification of important poxvirus-specific structural insertions. One insertion in the palm domain interacts with C-terminal residues of A20 and thus serves as the processivity factor-binding site. This is in strong contrast to all other family B polymerases that bind their co-factors at the C terminus of the thumb domain. The VACV E9 structure also permits rationalization of polymerase inhibitor resistance mutations when compared with the closely related eukaryotic polymerase delta-DNA complex.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Similar articles
-
Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.J Mol Biol. 2021 Jun 25;433(13):167009. doi: 10.1016/j.jmb.2021.167009. Epub 2021 Apr 24. J Mol Biol. 2021. PMID: 33901538
-
The vaccinia virus DNA polymerase and its processivity factor.Virus Res. 2017 Apr 15;234:193-206. doi: 10.1016/j.virusres.2017.01.027. Epub 2017 Feb 1. Virus Res. 2017. PMID: 28159613 Free PMC article. Review.
-
Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminal domain.PLoS Pathog. 2014 Mar 6;10(3):e1003978. doi: 10.1371/journal.ppat.1003978. eCollection 2014 Mar. PLoS Pathog. 2014. PMID: 24603707 Free PMC article.
-
Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase.J Biol Chem. 2006 Feb 10;281(6):3439-51. doi: 10.1074/jbc.M511239200. Epub 2005 Dec 1. J Biol Chem. 2006. PMID: 16326701
-
Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.Protein Sci. 2016 Dec;25(12):2113-2131. doi: 10.1002/pro.3058. Epub 2016 Nov 2. Protein Sci. 2016. PMID: 27684934 Free PMC article. Review.
Cited by
-
Poxvirus Recombination.Pathogens. 2022 Aug 9;11(8):896. doi: 10.3390/pathogens11080896. Pathogens. 2022. PMID: 36015016 Free PMC article. Review.
-
Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.PLoS Pathog. 2024 May 20;20(5):e1011652. doi: 10.1371/journal.ppat.1011652. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38768256 Free PMC article.
-
Efficient Method for Generating Point Mutations in the Vaccinia Virus Genome Using CRISPR/Cas9.Viruses. 2022 Jul 18;14(7):1559. doi: 10.3390/v14071559. Viruses. 2022. PMID: 35891539 Free PMC article.
-
Drug Development against Smallpox: Present and Future.Antimicrob Agents Chemother. 2020 Mar 24;64(4):e01683-19. doi: 10.1128/AAC.01683-19. Print 2020 Mar 24. Antimicrob Agents Chemother. 2020. PMID: 31932370 Free PMC article. Review.
-
Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.Sci Adv. 2024 Aug 23;10(34):eadl1150. doi: 10.1126/sciadv.adl1150. Epub 2024 Aug 21. Sci Adv. 2024. PMID: 39167653 Free PMC article.
References
-
- Moss, B. in Fields Virology 6th edn, Vol 2 (eds Fields, B. N., Knipe, D. M. & Howley, P. M.) 2129–2159 (Lippincott Williams & Wilkins, Philadelphia, 2013).
-
- Challberg MD, Englund PT. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. J. Biol. Chem. 1979;254:7812–7819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

