Trichloroethylene-induced alterations in DNA methylation were enriched in polycomb protein binding sites in effector/memory CD4+ T cells
- PMID: 29129997
- PMCID: PMC5676456
- DOI: 10.1093/eep/dvx013
Trichloroethylene-induced alterations in DNA methylation were enriched in polycomb protein binding sites in effector/memory CD4+ T cells
Abstract
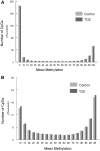
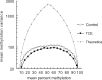
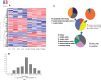
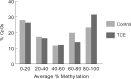
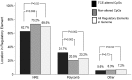
Exposure to industrial solvent and water pollutant trichloroethylene (TCE) can promote autoimmunity, and expand effector/memory (CD62L) CD4+ T cells. In order to better understand etiology reduced representation bisulfite sequencing was used to study how a 40-week exposure to TCE in drinking water altered methylation of ∼337 770 CpG sites across the entire genome of effector/memory CD4+ T cells from MRL+/+ mice. Regardless of TCE exposure, 62% of CpG sites in autosomal chromosomes were hypomethylated (0-15% methylation), and 25% were hypermethylated (85-100% methylation). In contrast, only 6% of the CpGs on the X chromosome were hypomethylated, and 51% had mid-range methylation levels. In terms of TCE impact, TCE altered (≥ 10%) the methylation of 233 CpG sites in effector/memory CD4+ T cells. Approximately 31.7% of these differentially methylated sites occurred in regions known to bind one or more Polycomb group (PcG) proteins, namely Ezh2, Suz12, Mtf2 or Jarid2. In comparison, only 23.3% of CpG sites not differentially methylated by TCE were found in PcG protein binding regions. Transcriptomics revealed that TCE altered the expression of ∼560 genes in the same effector/memory CD4+ T cells. At least 80% of the immune genes altered by TCE had binding sites for PcG proteins flanking their transcription start site, or were regulated by other transcription factors that were in turn ordered by PcG proteins at their own transcription start site. Thus, PcG proteins, and the differential methylation of their binding sites, may represent a new mechanism by which TCE could alter the function of effector/memory CD4+ T cells.
Keywords: DNA methylation; autoimmunity; immunotoxicity; polycomb proteins; trichloroethylene.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures
Similar articles
-
Continuous Developmental and Early Life Trichloroethylene Exposure Promoted DNA Methylation Alterations in Polycomb Protein Binding Sites in Effector/Memory CD4+ T Cells.Front Immunol. 2019 Aug 28;10:2016. doi: 10.3389/fimmu.2019.02016. eCollection 2019. Front Immunol. 2019. PMID: 31555266 Free PMC article.
-
Chronic exposure to trichloroethylene increases DNA methylation of the Ifng promoter in CD4+ T cells.Toxicol Lett. 2016 Oct 17;260:1-7. doi: 10.1016/j.toxlet.2016.08.017. Epub 2016 Aug 21. Toxicol Lett. 2016. PMID: 27553676 Free PMC article.
-
Chronic exposure to water pollutant trichloroethylene increased epigenetic drift in CD4(+) T cells.Epigenomics. 2016 May;8(5):633-49. doi: 10.2217/epi-2015-0018. Epub 2016 Apr 19. Epigenomics. 2016. PMID: 27092578 Free PMC article.
-
Trichloroethylene metabolite modulates DNA methylation-dependent gene expression in Th1-polarized CD4+ T cells from autoimmune-prone mice.Toxicol Sci. 2024 May 28;199(2):289-300. doi: 10.1093/toxsci/kfae032. Toxicol Sci. 2024. PMID: 38518092 Free PMC article.
-
Toxicological effects of trichloroethylene exposure on immune disorders.Immunopharmacol Immunotoxicol. 2017 Dec;39(6):305-317. doi: 10.1080/08923973.2017.1364262. Epub 2017 Aug 22. Immunopharmacol Immunotoxicol. 2017. PMID: 28828896 Review.
Cited by
-
Sex-Dependent Effects on Liver Inflammation and Gut Microbial Dysbiosis After Continuous Developmental Exposure to Trichloroethylene in Autoimmune-Prone Mice.Front Pharmacol. 2020 Oct 29;11:569008. doi: 10.3389/fphar.2020.569008. eCollection 2020. Front Pharmacol. 2020. PMID: 33250767 Free PMC article.
-
Opposing Actions of Developmental Trichloroethylene and High-Fat Diet Coexposure on Markers of Lipogenesis and Inflammation in Autoimmune-Prone Mice.Toxicol Sci. 2018 Jul 1;164(1):313-327. doi: 10.1093/toxsci/kfy091. Toxicol Sci. 2018. PMID: 29669109 Free PMC article.
-
Human exposure to trichloroethylene is associated with increased variability of blood DNA methylation that is enriched in genes and pathways related to autoimmune disease and cancer.Epigenetics. 2019 Nov;14(11):1112-1124. doi: 10.1080/15592294.2019.1633866. Epub 2019 Jun 26. Epigenetics. 2019. PMID: 31241004 Free PMC article.
-
Epigenetic underpinnings of developmental immunotoxicity and autoimmune disease.Curr Opin Toxicol. 2018 Aug;10:23-30. doi: 10.1016/j.cotox.2017.11.013. Epub 2017 Dec 1. Curr Opin Toxicol. 2018. PMID: 30613805 Free PMC article.
-
Differential Expression of miRNAs in Trichloroethene-Mediated Inflammatory/Autoimmune Response and Its Modulation by Sulforaphane: Delineating the Role of miRNA-21 and miRNA-690.Front Immunol. 2022 Mar 29;13:868539. doi: 10.3389/fimmu.2022.868539. eCollection 2022. Front Immunol. 2022. PMID: 35422807 Free PMC article.
References
-
- Kawakami N, Odoardi F, Ziemssen T, Bradl M, Ritter T, Neuhaus O, Lassmann H, Wekerle H, Flugel A.. Autoimmune CD4+ T cell memory: lifelong persistence of encephalitogenic T cell clones in healthy immune repertoires. J Immunol 2005;175:69–81. - PubMed
-
- Oling V, Reijonen H, Simell O, Knip M, Ilonen J.. Autoantigen-specific memory CD4+ T cells are prevalent early in progression to Type 1 diabetes. Cell Immunol 2012;273:133–9. - PubMed
-
- ATSDR: Toxicological Profile for Trichloroethylene, US Department of Health and Human Services, Agency for Toxic Substances and Disease Registry 2014. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

