Recurrent ubiquitin B silencing in gynecological cancers establishes dependence on ubiquitin C
- PMID: 29130934
- PMCID: PMC5707153
- DOI: 10.1172/JCI92914
Recurrent ubiquitin B silencing in gynecological cancers establishes dependence on ubiquitin C
Abstract
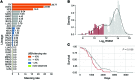
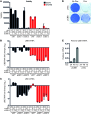
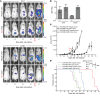
Transcriptional repression of ubiquitin B (UBB) is a cancer-subtype-specific alteration that occurs in a substantial population of patients with cancers of the female reproductive tract. UBB is 1 of 2 genes encoding for ubiquitin as a polyprotein consisting of multiple copies of ubiquitin monomers. Silencing of UBB reduces cellular UBB levels and results in an exquisite dependence on ubiquitin C (UBC), the second polyubiquitin gene. UBB is repressed in approximately 30% of high-grade serous ovarian cancer (HGSOC) patients and is a recurrent lesion in uterine carcinosarcoma and endometrial carcinoma. We identified ovarian tumor cell lines that retain UBB in a repressed state, used these cell lines to establish orthotopic ovarian tumors, and found that inducible expression of a UBC-targeting shRNA led to tumor regression, and substantial long-term survival benefit. Thus, we describe a recurrent cancer-specific lesion at the level of ubiquitin production. Moreover, these observations reveal the prognostic value of UBB repression and establish UBC as a promising therapeutic target for ovarian cancer patients with recurrent UBB silencing.
Keywords: Genetics; Oncology; Ubiquitin-proteosome system.
Conflict of interest statement
Figures





Comment in
-
Ubiquitin levels: the next target against gynecological cancers?J Clin Invest. 2017 Dec 1;127(12):4228-4230. doi: 10.1172/JCI98262. Epub 2017 Nov 13. J Clin Invest. 2017. PMID: 29130938 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

