JAK2-binding long noncoding RNA promotes breast cancer brain metastasis
- PMID: 29130936
- PMCID: PMC5707156
- DOI: 10.1172/JCI91553
JAK2-binding long noncoding RNA promotes breast cancer brain metastasis
Expression of concern in
-
Expression of Concern for JAK2-binding long noncoding RNA promotes breast cancer brain metastasis.J Clin Invest. 2025 Oct 1;135(19):e200302. doi: 10.1172/JCI200302. eCollection 2025 Oct 1. J Clin Invest. 2025. PMID: 41031879 Free PMC article. No abstract available.
Abstract
Conventional therapies for breast cancer brain metastases (BCBMs) have been largely ineffective because of chemoresistance and impermeability of the blood-brain barrier. A comprehensive understanding of the underlying mechanism that allows breast cancer cells to infiltrate the brain is necessary to circumvent treatment resistance of BCBMs. Here, we determined that expression of a long noncoding RNA (lncRNA) that we have named lncRNA associated with BCBM (Lnc-BM) is prognostic of the progression of brain metastasis in breast cancer patients. In preclinical murine models, elevated Lnc-BM expression drove BCBM, while depletion of Lnc-BM with nanoparticle-encapsulated siRNAs effectively treated BCBM. Lnc-BM increased JAK2 kinase activity to mediate oncostatin M- and IL-6-triggered STAT3 phosphorylation. In breast cancer cells, Lnc-BM promoted STAT3-dependent expression of ICAM1 and CCL2, which mediated vascular co-option and recruitment of macrophages in the brain, respectively. Recruited macrophages in turn produced oncostatin M and IL-6, thereby further activating the Lnc-BM/JAK2/STAT3 pathway and enhancing BCBM. Collectively, our results show that Lnc-BM and JAK2 promote BCBMs by mediating communication between breast cancer cells and the brain microenvironment. Moreover, these results suggest targeting Lnc-BM as a potential strategy for fighting this difficult disease.
Keywords: Breast cancer; Cell Biology; Oncology.
Conflict of interest statement
Figures
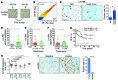
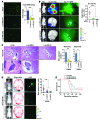


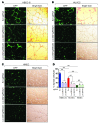




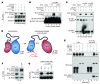

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

