Enhancing the precision of genetic lineage tracing using dual recombinases
- PMID: 29131159
- PMCID: PMC6913096
- DOI: 10.1038/nm.4437
Enhancing the precision of genetic lineage tracing using dual recombinases
Abstract
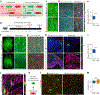
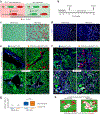
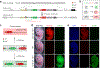
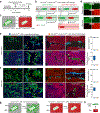
The Cre-loxP recombination system is the most widely used technology for in vivo tracing of stem or progenitor cell lineages. The precision of this genetic system largely depends on the specificity of Cre recombinase expression in targeted stem or progenitor cells. However, Cre expression in nontargeted cell types can complicate the interpretation of lineage-tracing studies and has caused controversy in many previous studies. Here we describe a new genetic lineage tracing system that incorporates the Dre-rox recombination system to enhance the precision of conventional Cre-loxP-mediated lineage tracing. The Dre-rox system permits rigorous control of Cre-loxP recombination in lineage tracing, effectively circumventing potential uncertainty of the cell-type specificity of Cre expression. Using this new system we investigated two topics of recent debates-the contribution of c-Kit+ cardiac stem cells to cardiomyocytes in the heart and the contribution of Sox9+ hepatic progenitor cells to hepatocytes in the liver. By overcoming the technical hurdle of nonspecific Cre-loxP-mediated recombination, this new technology provides more precise analysis of cell lineage and fate decisions and facilitates the in vivo study of stem and progenitor cell plasticity in disease and regeneration.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Snippert HJ et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

