An improved MS2 system for accurate reporting of the mRNA life cycle
- PMID: 29131164
- PMCID: PMC5843578
- DOI: 10.1038/nmeth.4502
An improved MS2 system for accurate reporting of the mRNA life cycle
Abstract
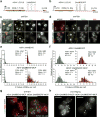
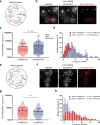
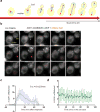
The MS2-MCP system enables researchers to image multiple steps of the mRNA life cycle with high temporal and spatial resolution. However, for short-lived mRNAs, the tight binding of the MS2 coat protein (MCP) to the MS2 binding sites (MBS) protects the RNA from being efficiently degraded, and this confounds the study of mRNA regulation. Here, we describe a reporter system (MBSV6) with reduced affinity for the MCP, which allows mRNA degradation while preserving single-molecule detection determined by single-molecule FISH (smFISH) or live imaging. Constitutive mRNAs (MDN1 and DOA1) and highly-regulated mRNAs (GAL1 and ASH1) endogenously tagged with MBSV6 in Saccharomyces cerevisiae degrade normally. As a result, short-lived mRNAs were imaged throughout their complete life cycle. The MBSV6 reporter revealed that, in contrast to previous findings, coordinated recruitment of mRNAs at specialized structures such as P-bodies during stress did not occur, and mRNA degradation was heterogeneously distributed in the cytoplasm.
Conflict of interest statement
Competing Financial Interest
The material in this manuscript is the subject of a provisional application to the US Patent and Trademark Office. It has not been licensed to any corporation, and the authors (E.T, M.V. and R.H.S) are the sole inventors.
Figures



References
-
- Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Molecular cell. 1998;2:437–445. doi:S1097-2765(00)80143-4 [pii] (1998) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

