Genetic and epigenetic regulation of major histocompatibility complex class I gene expression in bovine trophoblast cells
- PMID: 29131441
- PMCID: PMC5728445
- DOI: 10.1111/aji.12779
Genetic and epigenetic regulation of major histocompatibility complex class I gene expression in bovine trophoblast cells
Abstract
Problem: The regulatory mechanisms governing differential expression of classical major histocompatibility complex (MHC) class I (MHC-Ia) and non-classical MHC class I (MHC-Ib) genes are poorly understood.
Method of study: Quantitative reverse transcription- polymerase chain reaction (PCR) was used to compare the abundance of MHC-I transcripts and related transcription factors in peripheral blood mononuclear cells (PBMC) and placental trophoblast cells (PTC). Methylation of MHC-I CpG islands was detected by bisulfite treatment and next-generation sequencing. Demethylation of PBMC and PTC with 5'-aza-deoxycytidine was used to assess the role of methylation in gene regulation.
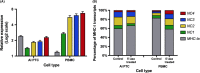
Results: MHC-I expression was higher in PBMC than PTC and was correlated with expression of IRF1, class II MHC transactivator (CIITA), and STAT1. The MHC-Ia genes and BoLA-NC1 were devoid of CpG methylation in PBMC and PTC. In contrast, CpG sites in the gene body of BoLA-NC2, -NC3, and -NC4 were highly methylated in PBMC but largely unmethylated in normal PTC and moderately methylated in somatic cell nuclear transfer PTC. In PBMC, demethylation resulted in upregulation of MHC-Ib by 2.8- to 6-fold, whereas MHC-Ia transcripts were elevated less than 2-fold.
Conclusion: DNA methylation regulates bovine MHC-Ib expression and is likely responsible for the different relative levels of MHC-Ib to MHC-Ia transcripts in PBMC and PTC.
Keywords: DNA methylation; bovine; non-classical MHC-I; transcription.
© 2017 The Authors. American Journal of Reproductive Immunology Published by John Wiley & Sons Ltd.
Figures





References
-
- Birch J, Codner G, Guzman E, Ellis SA. Genomic location and characterisation of nonclassical MHC class I genes in cattle. Immunogenetics. 2008;60:267‐273. - PubMed
-
- Davies CJ, Eldridge JA, Fisher PJ, Schlafer DH. Evidence for expression of both classical and non‐classical major histocompatibility complex class I genes in bovine trophoblast cells. Am J Reprod Immunol. 2006;55:188‐200. - PubMed
-
- Wick MJ, Ljunggren HG. Processing of bacterial antigens for peptide presentation on MHC class I molecules. Immunol Rev. 1999;172:153‐162. - PubMed
-
- Shastri N, Cardinaud S, Schwab SR, Serwold T, Kunisawa J. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen‐processing pathway. Immunol Rev. 2005;207:31‐41. - PubMed
-
- Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67‐74. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

