Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains
- PMID: 29131831
- PMCID: PMC5683605
- DOI: 10.1371/journal.pone.0187778
Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains
Abstract
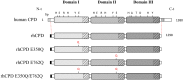
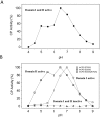
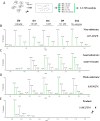
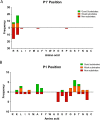
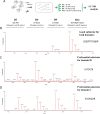
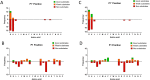
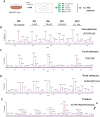
Metallocarboxypeptidase D (CPD) is a membrane-bound component of the trans-Golgi network that cycles to the cell surface through exocytic and endocytic pathways. Unlike other members of the metallocarboxypeptidase family, CPD is a multicatalytic enzyme with three carboxypeptidase-like domains, although only the first two domains are predicted to be enzymatically active. To investigate the enzymatic properties of each domain in human CPD, a critical active site Glu in domain I and/or II was mutated to Gln and the protein expressed, purified, and assayed with a wide variety of peptide substrates. CPD with all three domains intact displays >50% activity from pH 5.0 to 7.5 with a maximum at pH 6.5, as does CPD with mutation of domain I. In contrast, the domain II mutant displayed >50% activity from pH 6.5-7.5. CPD with mutations in both domains I and II was completely inactive towards all substrates and at all pH values. A quantitative peptidomics approach was used to compare the activities of CPD domains I and II towards a large number of peptides. CPD cleaved C-terminal Lys or Arg from a subset of the peptides. Most of the identified substrates of domain I contained C-terminal Arg, whereas comparable numbers of Lys- and Arg-containing peptides were substrates of domain II. We also report that some peptides with C-terminal basic residues were not cleaved by either domain I or II, showing the importance of the P1 position for CPD activity. Finally, the preference of domain I for C-terminal Arg was validated through molecular docking experiments. Together with the differences in pH optima, the different substrate specificities of CPD domains I and II allow the enzyme to perform distinct functions in the various locations within the cell.
Conflict of interest statement
Figures

Similar articles
-
Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain.Int J Mol Sci. 2020 Nov 18;21(22):8687. doi: 10.3390/ijms21228687. Int J Mol Sci. 2020. PMID: 33217972 Free PMC article.
-
Characterization of the enzymatic properties of the first and second domains of metallocarboxypeptidase D.J Biol Chem. 1999 Oct 8;274(41):28887-92. doi: 10.1074/jbc.274.41.28887. J Biol Chem. 1999. PMID: 10506132
-
Characterization of Drosophila carboxypeptidase D.J Biol Chem. 2002 Dec 20;277(51):49613-20. doi: 10.1074/jbc.M209652200. Epub 2002 Oct 21. J Biol Chem. 2002. PMID: 12393882
-
Purification and characterization of two serine carboxypeptidases from Aspergillus niger and their use in C-terminal sequencing of proteins and peptide synthesis.Appl Environ Microbiol. 1992 Jul;58(7):2144-52. doi: 10.1128/aem.58.7.2144-2152.1992. Appl Environ Microbiol. 1992. PMID: 1637154 Free PMC article.
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
Cited by
-
Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain.Int J Mol Sci. 2020 Nov 18;21(22):8687. doi: 10.3390/ijms21228687. Int J Mol Sci. 2020. PMID: 33217972 Free PMC article.
-
Enhanced Production of ECM Proteins for Pharmaceutical Applications Using Mammalian Cells and Sodium Heparin Supplementation.Pharmaceutics. 2022 Oct 8;14(10):2138. doi: 10.3390/pharmaceutics14102138. Pharmaceutics. 2022. PMID: 36297573 Free PMC article.
-
Biochemical and MALDI-TOF Mass Spectrometric Characterization of a Novel Native and Recombinant Cystine Knot Miniprotein from Solanum tuberosum subsp. andigenum cv. Churqueña.Int J Mol Sci. 2018 Feb 28;19(3):678. doi: 10.3390/ijms19030678. Int J Mol Sci. 2018. PMID: 29495576 Free PMC article.
-
Inactive metallopeptidase homologs: the secret lives of pseudopeptidases.Front Mol Biosci. 2024 Jul 10;11:1436917. doi: 10.3389/fmolb.2024.1436917. eCollection 2024. Front Mol Biosci. 2024. PMID: 39050735 Free PMC article. Review.
-
Complement activity and autophagy are dysregulated in the lungs of patients with nonresolvable COVID-19 requiring lung transplantation.Respir Res. 2025 Feb 27;26(1):68. doi: 10.1186/s12931-025-03152-6. Respir Res. 2025. PMID: 40016722 Free PMC article.
References
-
- Arolas JL, Vendrell J, Aviles FX, Fricker LD (2007) Metallocarboxypeptidases: emerging drug targets in biomedicine. Curr Pharm Des 13: 349–366. - PubMed
-
- Gomis-Ruth FX (2008) Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol 43: 319–345. doi: 10.1080/10409230802376375 - DOI - PubMed
-
- Rawlings ND, Waller M, Barrett AJ, Bateman A (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 42: D503–509. doi: 10.1093/nar/gkt953 - DOI - PMC - PubMed
-
- Lyons PJ, Fricker LD (2011) Carboxypeptidase O Is a Glycosylphosphatidylinositol-anchored Intestinal Peptidase with Acidic Amino Acid Specificity. Journal of Biological Chemistry 286: 39023–39032. doi: 10.1074/jbc.M111.265819 - DOI - PMC - PubMed
-
- Reznik SE, Fricker LD (2001) Carboxypeptidases from A to Z: implications in embryonic development and Wnt binding. Cellular and Molecular Life Sciences 58: 1790–1804. doi: 10.1007/PL00000819 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

