Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains
- PMID: 29131831
- PMCID: PMC5683605
- DOI: 10.1371/journal.pone.0187778
Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains
Abstract
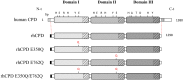
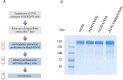
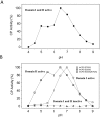
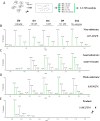
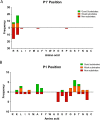
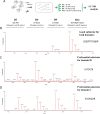
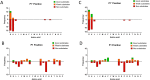
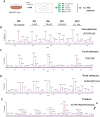
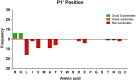
Metallocarboxypeptidase D (CPD) is a membrane-bound component of the trans-Golgi network that cycles to the cell surface through exocytic and endocytic pathways. Unlike other members of the metallocarboxypeptidase family, CPD is a multicatalytic enzyme with three carboxypeptidase-like domains, although only the first two domains are predicted to be enzymatically active. To investigate the enzymatic properties of each domain in human CPD, a critical active site Glu in domain I and/or II was mutated to Gln and the protein expressed, purified, and assayed with a wide variety of peptide substrates. CPD with all three domains intact displays >50% activity from pH 5.0 to 7.5 with a maximum at pH 6.5, as does CPD with mutation of domain I. In contrast, the domain II mutant displayed >50% activity from pH 6.5-7.5. CPD with mutations in both domains I and II was completely inactive towards all substrates and at all pH values. A quantitative peptidomics approach was used to compare the activities of CPD domains I and II towards a large number of peptides. CPD cleaved C-terminal Lys or Arg from a subset of the peptides. Most of the identified substrates of domain I contained C-terminal Arg, whereas comparable numbers of Lys- and Arg-containing peptides were substrates of domain II. We also report that some peptides with C-terminal basic residues were not cleaved by either domain I or II, showing the importance of the P1 position for CPD activity. Finally, the preference of domain I for C-terminal Arg was validated through molecular docking experiments. Together with the differences in pH optima, the different substrate specificities of CPD domains I and II allow the enzyme to perform distinct functions in the various locations within the cell.
Conflict of interest statement
Figures

References
-
- Arolas JL, Vendrell J, Aviles FX, Fricker LD (2007) Metallocarboxypeptidases: emerging drug targets in biomedicine. Curr Pharm Des 13: 349–366. - PubMed
-
- Gomis-Ruth FX (2008) Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol 43: 319–345. doi: 10.1080/10409230802376375 - DOI - PubMed
-
- Rawlings ND, Waller M, Barrett AJ, Bateman A (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 42: D503–509. doi: 10.1093/nar/gkt953 - DOI - PMC - PubMed
-
- Lyons PJ, Fricker LD (2011) Carboxypeptidase O Is a Glycosylphosphatidylinositol-anchored Intestinal Peptidase with Acidic Amino Acid Specificity. Journal of Biological Chemistry 286: 39023–39032. doi: 10.1074/jbc.M111.265819 - DOI - PMC - PubMed
-
- Reznik SE, Fricker LD (2001) Carboxypeptidases from A to Z: implications in embryonic development and Wnt binding. Cellular and Molecular Life Sciences 58: 1790–1804. doi: 10.1007/PL00000819 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

