A functional connection between dyskerin and energy metabolism
- PMID: 29132127
- PMCID: PMC5684492
- DOI: 10.1016/j.redox.2017.11.003
A functional connection between dyskerin and energy metabolism
Abstract
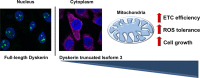
The human DKC1 gene encodes dyskerin, an evolutionarily conserved nuclear protein whose overexpression represents a common trait of many types of aggressive sporadic cancers. As a crucial component of the nuclear H/ACA snoRNP complexes, dyskerin is involved in a variety of essential processes, including telomere maintenance, splicing efficiency, ribosome biogenesis, snoRNAs stabilization and stress response. Although multiple minor dyskerin splicing isoforms have been identified, their functions remain to be defined. Considering that low-abundance splice variants could contribute to the wide functional repertoire attributed to dyskerin, possibly having more specialized tasks or playing significant roles in changing cell status, we investigated in more detail the biological roles of a truncated dyskerin isoform that lacks the C-terminal nuclear localization signal and shows a prevalent cytoplasmic localization. Here we show that this dyskerin variant can boost energy metabolism and improve respiration, ultimately conferring a ROS adaptive response and a growth advantage to cells. These results reveal an unexpected involvement of DKC1 in energy metabolism, highlighting a previously underscored role in the regulation of metabolic cell homeostasis.
Keywords: DKC1; Energy metabolism; Mitochondria; PRDX-2; ROS signaling.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Heiss N.S., Knight S.W., Vulliamy T.J., Klauck S.M., Wiemann S., Mason P.J., Poustka A., Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. - PubMed
-
- Cohen S.B., Graham M.E., Lovrecz G.O., Bache N., Robinson P.J., Reddel R.R. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. - PubMed
-
- Dokal I. Dyskeratosis congenita. Hematol. Am. Soc. Hematol. Educ. Prog. 2011:480–486. - PubMed
-
- Angrisani A., Vicidomini R., Turano M., Furia M. Human dyskerin: beyond telomeres. Biol. Chem. 2014;395:593–610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

