Effect of two non-synonymous ecto-5'-nucleotidase variants on the genetic architecture of inosine 5'-monophosphate (IMP) and its degradation products in Japanese Black beef
- PMID: 29132308
- PMCID: PMC5683534
- DOI: 10.1186/s12864-017-4275-4
Effect of two non-synonymous ecto-5'-nucleotidase variants on the genetic architecture of inosine 5'-monophosphate (IMP) and its degradation products in Japanese Black beef
Abstract
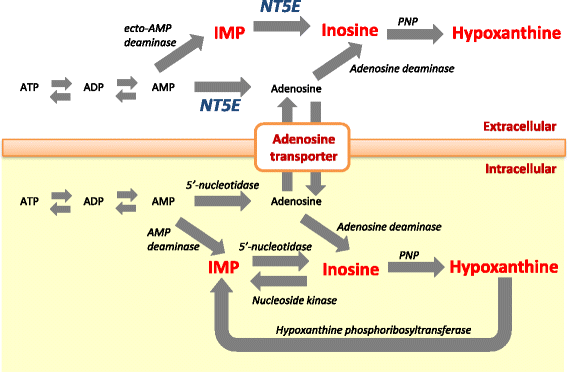
Background: Umami is a Japanese term for the fifth basic taste and is an important sensory property of beef palatability. Inosine 5'-monophosphate (IMP) contributes to umami taste in beef. Thus, the overall change in concentration of IMP and its degradation products can potentially affect the beef palatability. In this study, we investigated the genetic architecture of IMP and its degradation products in Japanese Black beef. First, we performed genome-wide association study (GWAS), candidate gene analysis, and functional analysis to detect the causal variants that affect IMP, inosine, and hypoxanthine. Second, we evaluated the allele frequencies in the different breeds, the contribution of genetic variance, and the effect on other economical traits using the detected variants.
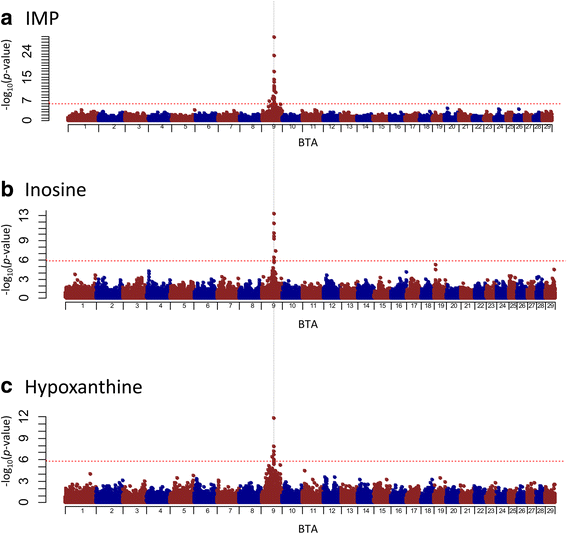
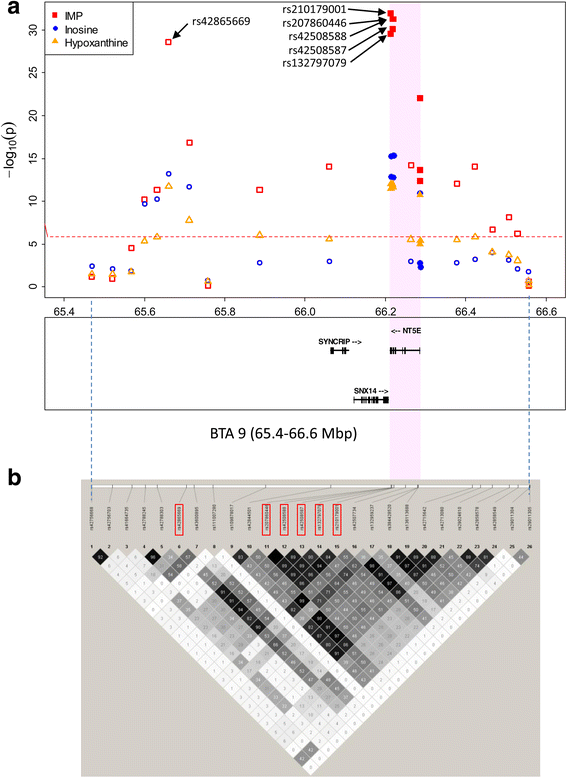
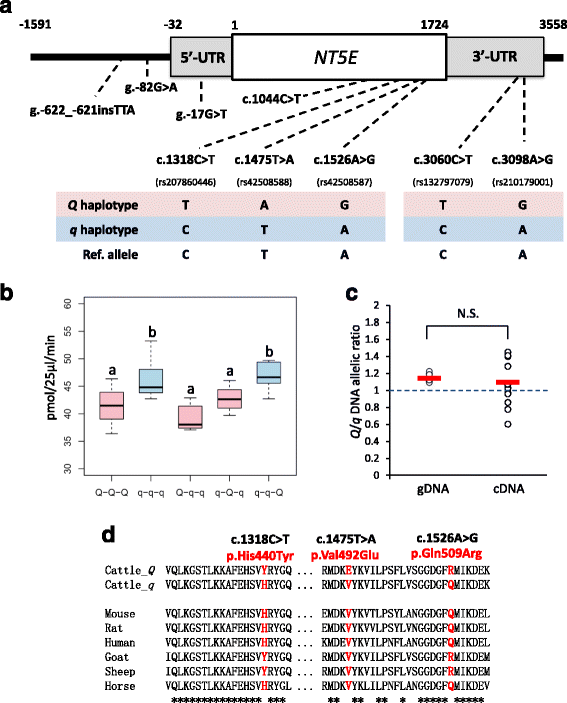
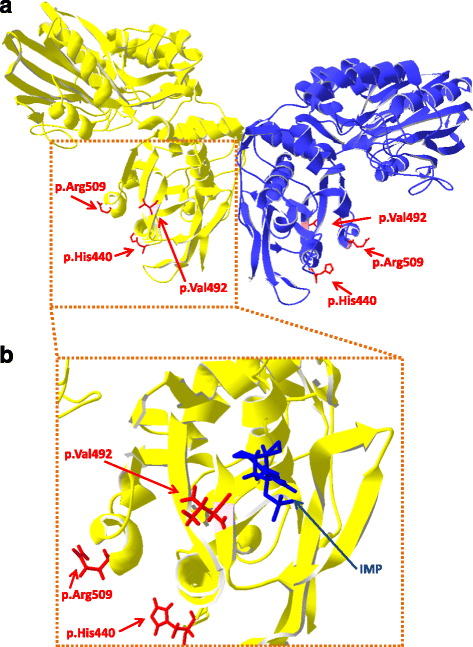
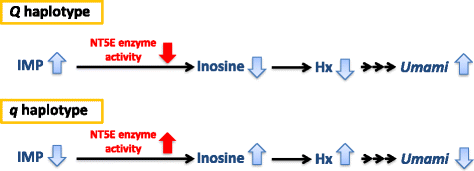
Results: A total of 574 Japanese Black cattle were genotyped using the Illumina BovineSNP50 BeadChip and were then used for GWAS. The results of GWAS showed that the genome-wide significant single nucleotide polymorphisms (SNPs) on BTA9 were detected for IMP, inosine, and hypoxanthine. The ecto-5'-nucleotidase (NT5E) gene, which encodes the enzyme NT5E for the extracellular degradation of IMP to inosine, was located near the significant region on BTA9. The results of candidate gene analysis and functional analysis showed that two non-synonymous SNPs (c.1318C > T and c.1475 T > A) in NT5E affected the amount of IMP and its degradation products in beef by regulating the enzymatic activity of NT5E. The Q haplotype showed a positive effect on IMP and a negative effect on the enzymatic activity of NT5E in IMP degradation. The two SNPs were under perfect linkage disequilibrium in five different breeds, and different haplotype frequencies were seen among breeds. The two SNPs contribute to about half of the total genetic variance in IMP, and the results of genetic relationship between IMP and its degradation products showed that NT5E affected the overall concentration balance of IMP and its degradation products. In addition, the SNPs in NT5E did not have an unfavorable effect on the other economical traits.
Conclusion: Based on all the above findings taken together, two non-synonymous SNPs in NT5E would be useful for improving IMP and its degradation products by marker-assisted selection in Japanese Black cattle.
Keywords: GWAS; IMP; Japanese Black cattle; Meat quality; NT5E.
Conflict of interest statement
Ethics approval and consent to participate
For GWAS and positional candidate gene analysis, Animal Care and Use Committee approval was not required. This was because the data were collected from beef cattle shipped to a meat processing plant in Yamagata Prefecture, Japan. For functional analysis and genotype and haplotype frequencies analyses among breeds, all procedures involving samples followed the Guidelines for the Animal Care and Use of Laboratory Animals established by National Livestock Breeding Center (NLBC) in Japan, and this research was approved by the laboratory animals committee on the NLBC.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests to National Livestock Breeding Center.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Gill JL, Matika O, Williams JL, Worton H, Wiener P, Bishop SC. Consistency statistics and genetic parameter for taste panel associated meat quality traits and their relationship with carcass quality traits in a commercial population of Angus-sired beef cattle. Animal. 2010;4:1–8. doi: 10.1017/S1751731109990905. - DOI - PubMed
-
- Pegg RB, Shahidi F. Heat effects on meat/warmed-oven flavour. In: Jensen WK, Devine C, Dikeman M, editors. Encyclopedia of meat sciences Oxford: Elsevier ltd. 2004. pp. 592–599.
-
- Nishimura T, Rhue MR, Okitani A, Kato H. Components contributing to the improvement of meat taste during storage. Agric Biol Chem. 1988;52:2323–2330.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

