Genomic analysis of endemic clones of toxigenic and non-toxigenic Corynebacterium diphtheriae in Belarus during and after the major epidemic in 1990s
- PMID: 29132312
- PMCID: PMC5683216
- DOI: 10.1186/s12864-017-4276-3
Genomic analysis of endemic clones of toxigenic and non-toxigenic Corynebacterium diphtheriae in Belarus during and after the major epidemic in 1990s
Abstract
Background: Diphtheria remains a major public health concern with multiple recent outbreaks around the world. Moreover, invasive non-toxigenic strains have emerged globally causing severe infections. A diphtheria epidemic in the former Soviet Union in the 1990s resulted in ~5000 deaths. In this study, we analysed the genome sequences of a collection of 93 C. diphtheriae strains collected during and after this outbreak (1996 - 2014) in a former Soviet State, Belarus to understand the evolutionary dynamics and virulence capacities of these strains.
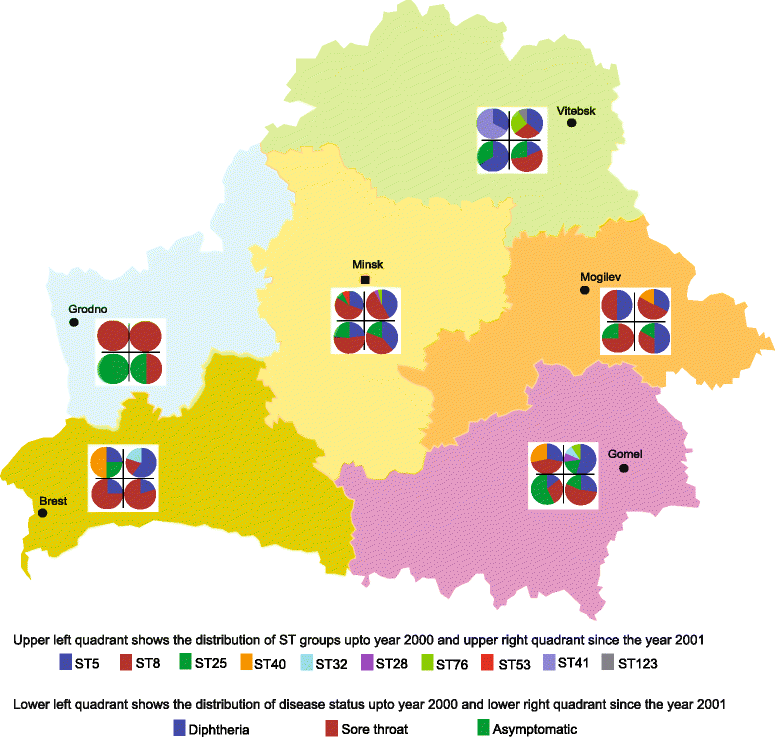
Results: C. diphtheriae strains from Belarus belong to ten sequence types (STs). Two major clones, non-toxigenic ST5 and toxigenic ST8, encompassed 76% of the isolates that are associated with sore throat and diphtheria in patients, respectively. Core genomic diversity is limited within outbreak-associated ST8 with relatively higher mutation rates (8.9 × 10-7 substitutions per strain per year) than ST5 (5.6 × 10-7 substitutions per strain per year) where most of the diversity was introduced by recombination. A variation in the virulence gene repertoire including the presence of tox gene is likely responsible for pathogenic differences between different strains. However, strains with similar virulence potential can cause disease in some individuals and remain asymptomatic in others. Eight synonymous single nucleotide polymorphisms were observed between the tox genes of the vaccine strain PW8 and other toxigenic strains of ST8, ST25, ST28, ST41 and non-toxigenic tox gene-bearing (NTTB) ST40 strains. A single nucleotide deletion at position 52 in the tox gene resulted in the frameshift in ST40 isolates, converting them into NTTB strains.
Conclusions: Non-toxigenic C. diphtheriae ST5 and toxigenic ST8 strains have been endemic in Belarus both during and after the epidemic in 1990s. A high vaccine coverage has effectively controlled diphtheria in Belarus; however, non-toxigenic strains continue to circulate in the population. Recombination is an important evolutionary force in shaping the genomic diversity in C. diphtheriae. However, the relative role of recombination and mutations in diversification varies between different clones.
Keywords: Corynebacterium diphtheriae; Diphtheria; Endemic; Epidemic; Non-toxigenic; Sore throat; Toxigenic; Vaccine; Virulence.
Conflict of interest statement
Ethics approval and consent to participate
The authors declare that this study does not involve any human subjects, human material, human data, animals, animal material, plants or any plant material. Therefore, such an ethical approval is not applicable to this study.
Consent for publication
This manuscript does not include any details, images, or videos relating to an individual person. Therefore, consent for publication is not applicable to this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Genomic Analysis of Corynebacterium diphtheriae Strains Isolated in the Years 2007-2022 with a Report on the Identification of the First Non-Toxigenic tox Gene-Bearing Strain in Poland.Int J Mol Sci. 2023 Feb 27;24(5):4612. doi: 10.3390/ijms24054612. Int J Mol Sci. 2023. PMID: 36902043 Free PMC article.
-
Changes in MLST profiles and biotypes of Corynebacterium diphtheriae isolates from the diphtheria outbreak period to the period of invasive infections caused by nontoxigenic strains in Poland (1950-2016).BMC Infect Dis. 2018 Mar 9;18(1):121. doi: 10.1186/s12879-018-3020-1. BMC Infect Dis. 2018. PMID: 29523087 Free PMC article.
-
Evolution, epidemiology and diversity of Corynebacterium diphtheriae: New perspectives on an old foe.Infect Genet Evol. 2016 Sep;43:364-70. doi: 10.1016/j.meegid.2016.06.024. Epub 2016 Jun 9. Infect Genet Evol. 2016. PMID: 27291708 Review.
-
Molecular epidemiology of C. diphtheriae strains during different phases of the diphtheria epidemic in Belarus.BMC Infect Dis. 2006 Aug 15;6:129. doi: 10.1186/1471-2334-6-129. BMC Infect Dis. 2006. PMID: 16911772 Free PMC article.
-
Corynebacterium diphtheriae: genome diversity, population structure and genotyping perspectives.Infect Genet Evol. 2009 Jan;9(1):1-15. doi: 10.1016/j.meegid.2008.09.011. Epub 2008 Oct 19. Infect Genet Evol. 2009. PMID: 19007916 Review.
Cited by
-
Phenotypic and Genotypic Correlates of Penicillin Susceptibility in Nontoxigenic Corynebacterium diphtheriae, British Columbia, Canada, 2015-2018.Emerg Infect Dis. 2020 Jan;26(1):97-103. doi: 10.3201/eid2601.191241. Emerg Infect Dis. 2020. PMID: 31855139 Free PMC article.
-
Genomic analyses reveal two distinct lineages of Corynebacterium ulcerans strains.New Microbes New Infect. 2018 May 25;25:7-13. doi: 10.1016/j.nmni.2018.05.005. eCollection 2018 Sep. New Microbes New Infect. 2018. PMID: 29997890 Free PMC article.
-
Genomic Analysis of Corynebacterium diphtheriae Strains Isolated in the Years 2007-2022 with a Report on the Identification of the First Non-Toxigenic tox Gene-Bearing Strain in Poland.Int J Mol Sci. 2023 Feb 27;24(5):4612. doi: 10.3390/ijms24054612. Int J Mol Sci. 2023. PMID: 36902043 Free PMC article.
-
Distinct Genomic Features Characterize Two Clades of Corynebacterium diphtheriae: Proposal of Corynebacterium diphtheriae Subsp. diphtheriae Subsp. nov. and Corynebacterium diphtheriae Subsp. lausannense Subsp. nov.Front Microbiol. 2018 Aug 17;9:1743. doi: 10.3389/fmicb.2018.01743. eCollection 2018. Front Microbiol. 2018. PMID: 30174653 Free PMC article.
-
Genomic analysis of a novel nontoxigenic Corynebacterium diphtheriae strain isolated from a cancer patient.New Microbes New Infect. 2019 Apr 9;30:100544. doi: 10.1016/j.nmni.2019.100544. eCollection 2019 Jul. New Microbes New Infect. 2019. PMID: 31061711 Free PMC article.
References
-
- Malito E, Rappouli R. History of diphtheria vaccine development. In: Burkovski A, editor. Corynebacterium diphtheriae and related toxigenic species. Heidelberg: Springer; 2014. p. 225–38.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

