Development of amplicon deep sequencing markers and data analysis pipeline for genotyping multi-clonal malaria infections
- PMID: 29132317
- PMCID: PMC5682641
- DOI: 10.1186/s12864-017-4260-y
Development of amplicon deep sequencing markers and data analysis pipeline for genotyping multi-clonal malaria infections
Abstract
Background: Amplicon deep sequencing permits sensitive detection of minority clones and improves discriminatory power for genotyping multi-clone Plasmodium falciparum infections. New amplicon sequencing and data analysis protocols are needed for genotyping in epidemiological studies and drug efficacy trials of P. falciparum.
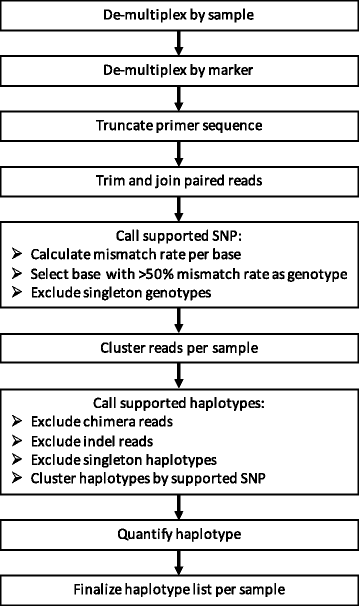
Methods: Targeted sequencing of molecular marker csp and novel marker cpmp was conducted in duplicate on mixtures of parasite culture strains and 37 field samples. A protocol allowing to multiplex up to 384 samples in a single sequencing run was applied. Software "HaplotypR" was developed for data analysis.
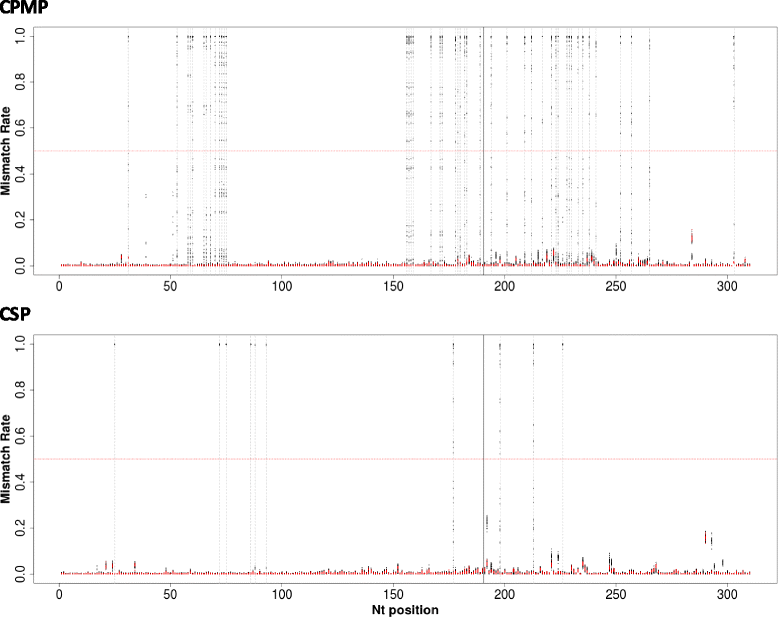
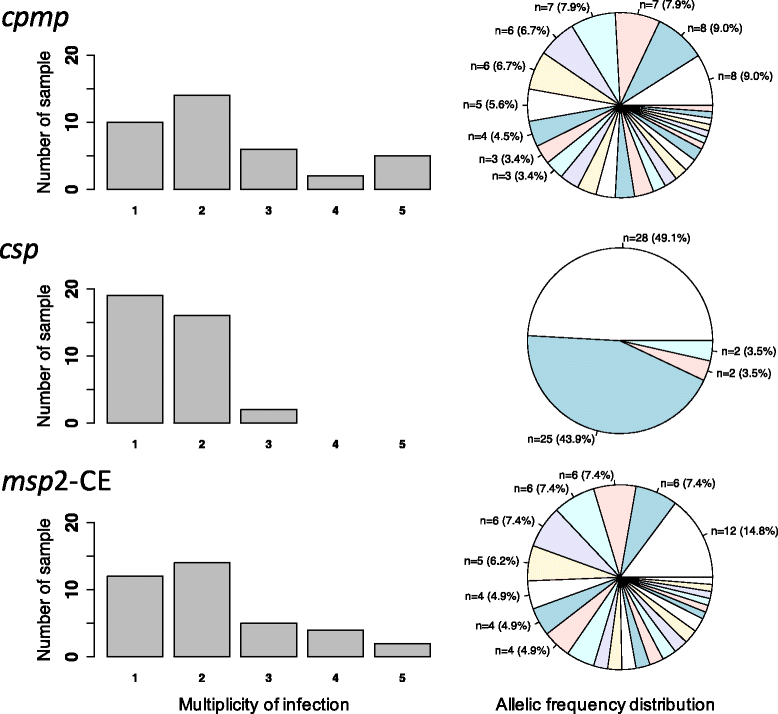
Results: Cpmp was highly diverse (He = 0.96) in contrast to csp (He = 0.57). Minority clones were robustly detected if their frequency was >1%. False haplotype calls owing to sequencing errors were observed below that threshold.
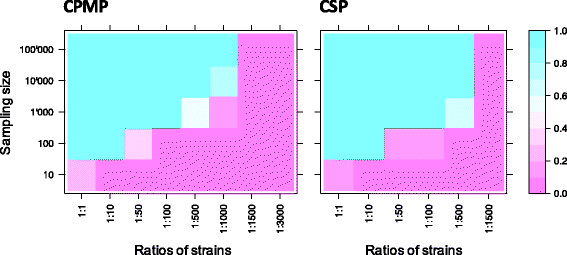
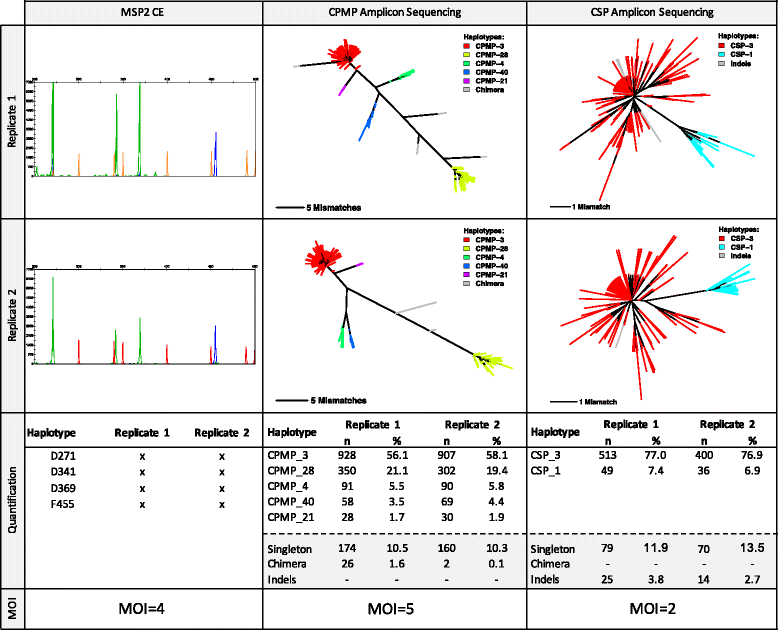
Conclusions: To reliably detect haplotypes at very low frequencies, experiments are best performed in duplicate and should aim for coverage of >10'000 reads/amplicon. When compared to length polymorphic marker msp2, highly multiplexed amplicon sequencing displayed greater sensitivity in detecting minority clones.
Keywords: Amplicon sequencing; HaplotypR software; Haplotype clustering; Malaria; Multi-clone infections; Plasmodium falciparum; SNP; cpmp; csp; msp2.
Conflict of interest statement
Ethics approval and consent to participate
Ethical clearance was obtained from PNG Institute of Medical Research Institutional Review Board (IRB 07.20) and PNG Medical Advisory Committee (07.34). Informed written consent was obtained from all parents or guardians prior to recruitment of each child. No medical records were used for this study.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- World Health Organization . Methods and techniques for clinical trials on antimalarial drug efficacy: genotyping to identify parasite populations. 2008.
-
- Falk N, Maire N, Sama W, Owusu-Agyei S, Smith T, Beck H-P, et al. Comparison of PCR-RFLP and Genescan-based genotyping for analyzing infection dynamics of plasmodium falciparum. Am J Trop Med Hyg. 2006;74:944–950. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

