Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma
- PMID: 29132337
- PMCID: PMC5683385
- DOI: 10.1186/s12885-017-3729-z
Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma
Abstract
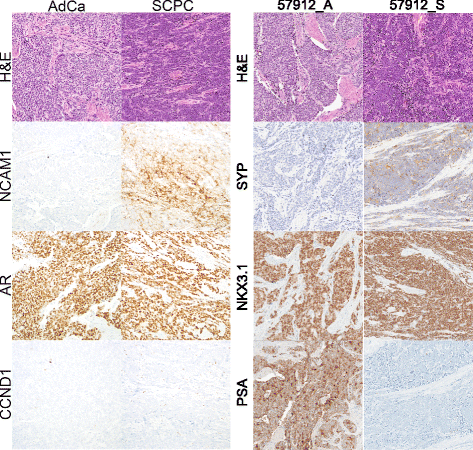
Background: Neuroendocrine prostate cancer (NEPC) may be rising in prevalence as patients with advanced prostate cancer potentially develop resistance to contemporary anti-androgen treatment through a neuroendocrine phenotype. While prior studies comparing NEPC and prostatic adenocarcinoma have identified important candidates for targeted therapy, most have relied on few NEPC patients due to disease rarity, resulting in thousands of differentially expressed genes collectively and offering an opportunity for meta-analysis. Moreover, past studies have focused on prototypical NEPC samples with classic immunohistochemistry profiles, whereas there is increasing recognition of atypical phenotypes. In the primary setting, small cell prostatic carcinoma (SCPC) is frequently admixed with adenocarcinomas that may be clonally related, and a minority of SCPCs express markers typical of prostatic adenocarcinoma while rare cases do not express neuroendocrine markers. We derived a meta-signature of prototypical high-grade NEPC, then applied it to develop a classifier of primary SCPC incorporating disease heterogeneity.
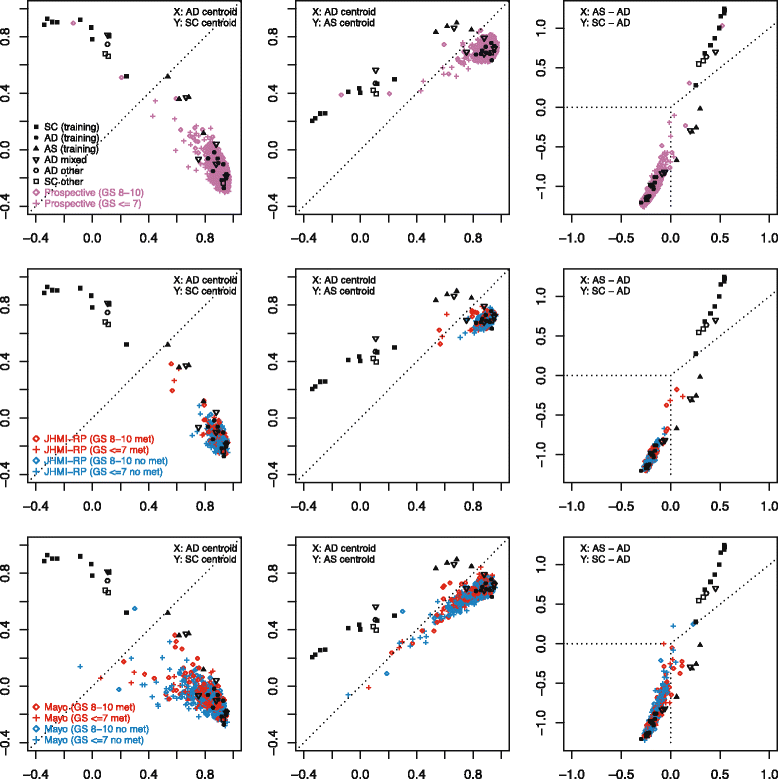
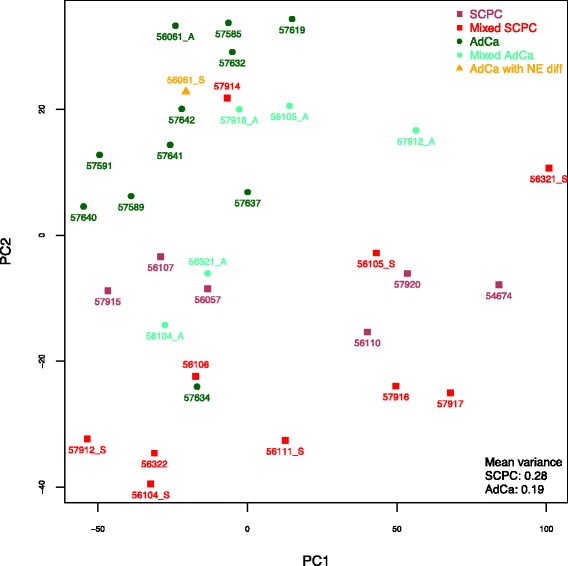
Methods: Prototypical NEPC samples from 15 patients across 6 frozen tissue microarray datasets were assessed for genes with consistent outlier expression relative to adenocarcinomas. Resulting genes were used to determine subgroups of primary SCPCs (N=16) and high-grade adenocarcinomas (N=16) profiled by exon arrays using formalin-fixed paraffin-embedded (FFPE) material from our institutional archives. A subgroup classifier was developed using differential expression for feature selection, and applied to radical prostatectomy cohorts.
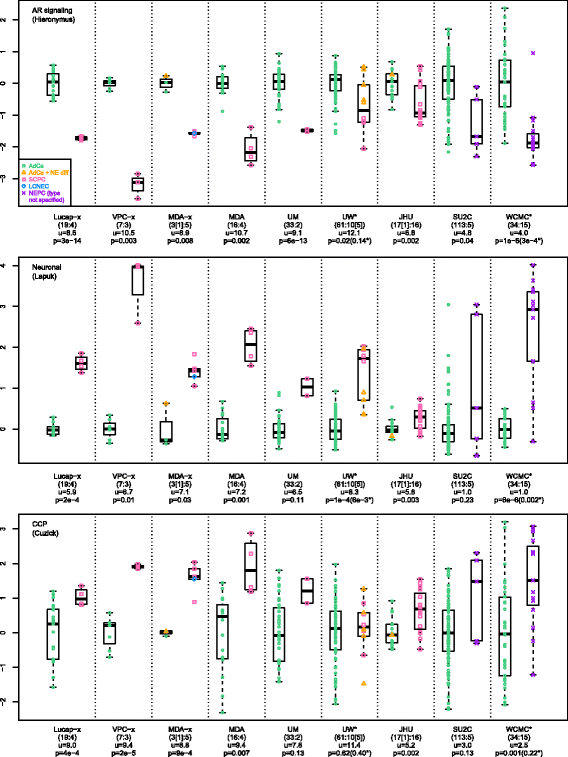
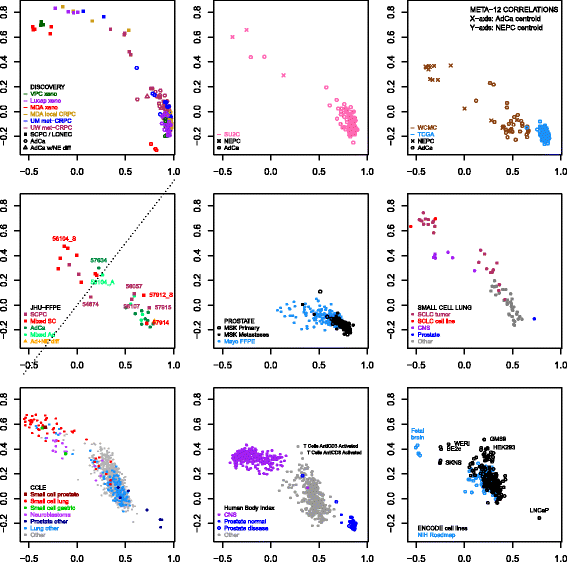
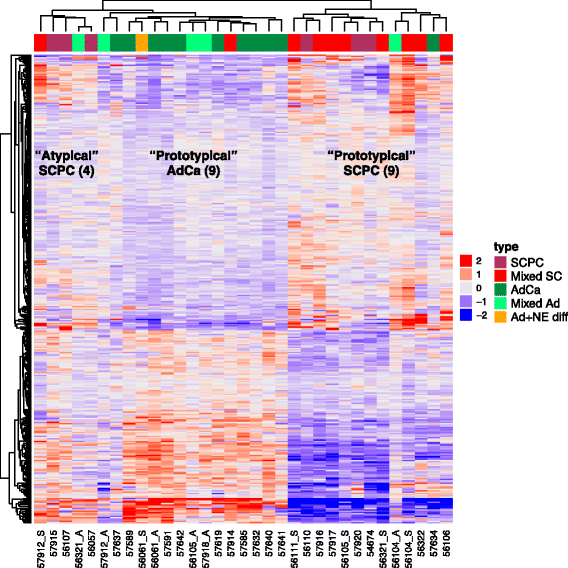
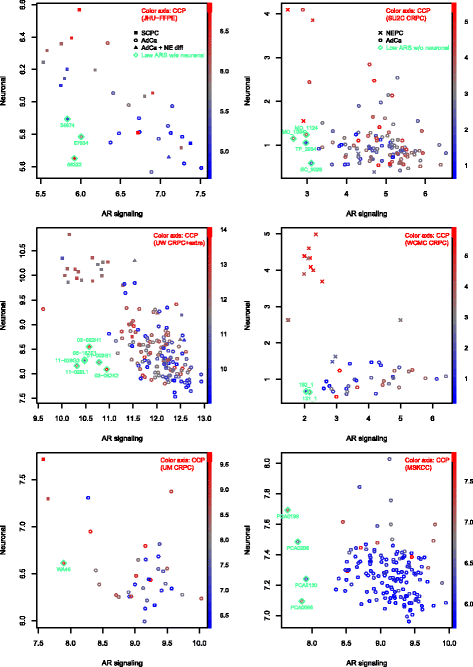
Results: Sixty nine and 375 genes demonstrated consistent outlier expression in at least 80% and 60% of NEPC patients, with close resemblance in expression between NEPC and small cell lung cancer. Clustering by these genes generated 3 subgroups among primary samples from our institution. Nearest centroid classification based on the predominant phenotype from each subgroup (9 prototypical SCPCs, 9 prototypical adenocarcinomas, and 4 atypical SCPCs) achieved a 4.5% error rate by leave-one-out cross-validation. The classifier identified SCPC-like expression in 40% (2/5) of mixed adenocarcinomas and 0.3-0.6% of adenocarcinomas from prospective (4/2293) and retrospective (2/355) radical prostatectomy cohorts, where both SCPC-like retrospective cases subsequently developed metastases.
Conclusions: Meta-analysis generates a robust signature of prototypical high-grade NEPC, and may facilitate development of a primary SCPC classifier based on FFPE material with potential prognostic implications.
Keywords: FFPE; Gene signature; Meta-analysis; Mixed prostatic adenocarcinoma; Nearest centroid classifier; Neuroendocrine prostate cancer; Small cell carcinoma.
Conflict of interest statement
Ethics approval and consent to participate
Informed consent to use the tissue samples in this study was waived by the John Hopkins School of Medicine Institutional Review Board.
Consent for publication
Not applicable.
Competing interests
JL, MA, NE and ED are employees of GenomeDx Biosciences; TLL has received research funding from GenomeDx. HKT declares no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Wang HT, et al. Neuroendocrine prostate cancer (nepc) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of nepc and survival from nepc diagnosis-a systematic review and pooled analysis. J Clin Oncol. 2014;32(30):3383–3390. doi: 10.1200/JCO.2013.54.3553. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

