Aptamer selection and applications for breast cancer diagnostics and therapy
- PMID: 29132385
- PMCID: PMC5683342
- DOI: 10.1186/s12951-017-0311-4
Aptamer selection and applications for breast cancer diagnostics and therapy
Abstract
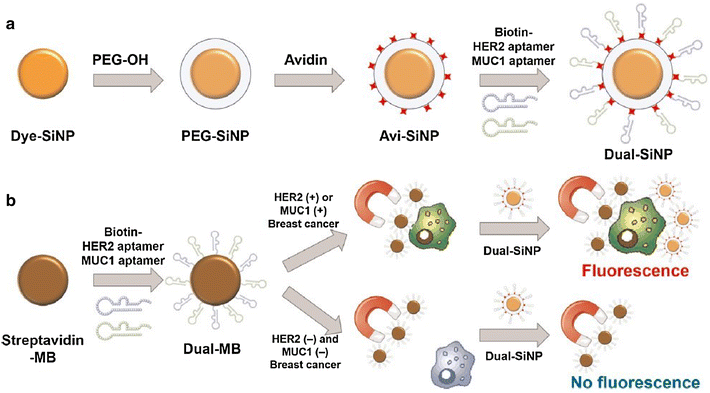
Aptamers are short non-coding, single-stranded oligonucleotides (RNA or DNA) developed through Systematic Evolution of Ligands by Exponential enrichment (SELEX) in vitro. Similar to antibodies, aptamers can bind to specific targets with high affinity, and are considered promising therapeutic agents as they have several advantages over antibodies, including high specificity, stability, and non-immunogenicity. Furthermore, aptamers can be produced at a low cost and easily modified, and are, therefore, called "chemical antibodies". In the past years, a variety of aptamers specifically bound to both breast cancer biomarkers and cells had been selected. Besides, taking advantage of nanomaterials, there were a number of aptamer-nanomaterial conjugates been developed and widely investigated for diagnostics and targeted therapy of breast cancer. In this short review, we first present a systematical review of various aptamer selection methods. Then, various aptamer-based diagnostic and therapeutic strategies of breast cancer were provided. Finally, the current problems, challenges, and future perspectives in the field were thoroughly discussed.
Keywords: Aptamer; Breast cancer; Diagnosis; SELEX; Targeted therapy.
Figures
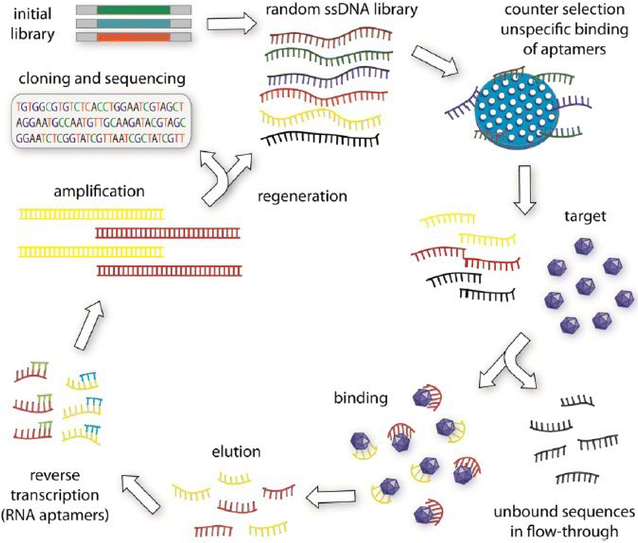
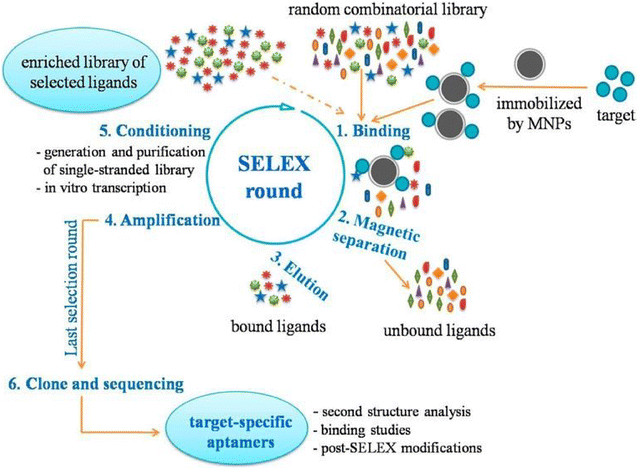

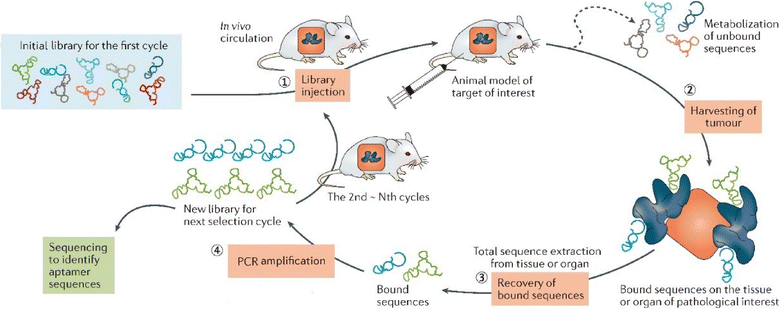


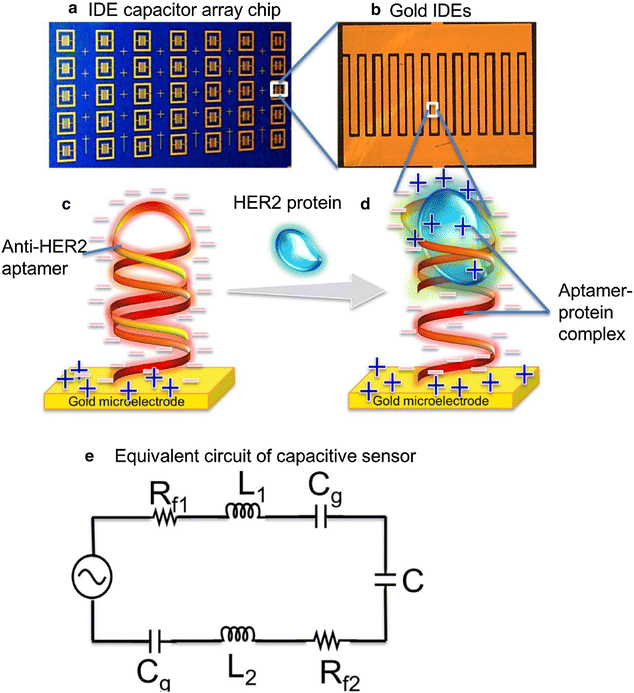


References
Publication types
MeSH terms
Substances
Grants and funding
- 2017YFA0205300/the National Key Research and Development Program of China
- SKLOD2017OF04/Open Funding of State Key Laboratory of Oral Diseases
- 2016T90403/China Postdoctoral Science Foundation
- (2013) 448/the Economical Forest Cultivation and Utilization of 2011 Collaborative Innovation Center in Hunan Province
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

