A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication
- PMID: 29132390
- PMCID: PMC5683209
- DOI: 10.1186/s12964-017-0201-2
A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication
Abstract
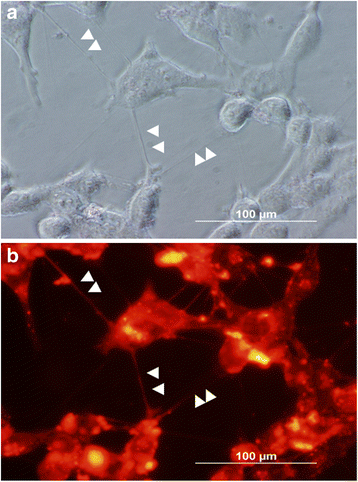
Background: Tunneling nanotubes (TNTs) are naturally-occurring filamentous actin-based membranous extensions that form across a wide spectrum of mammalian cell types to facilitate long-range intercellular communication. Valid assays are needed to accurately assess the downstream effects of TNT-mediated transfer of cellular signals in vitro. We recently reported a modified transwell assay system designed to test the effects of intercellular transfer of a therapeutic oncolytic virus, and viral-activated drugs, between cells via TNTs. The objective of the current study was to demonstrate validation of this in vitro approach as a new method for effectively excluding diffusible forms of long- and close-range intercellular transfer of intracytoplasmic cargo, including exosomes/microvesicles and gap junctions in order to isolate TNT-selective cell communication.
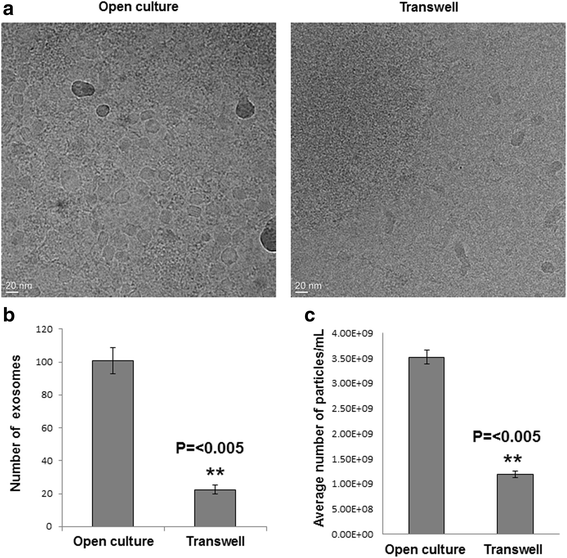
Methods: We designed several steps to effectively reduce or eliminate diffusion and long-range transfer via these extracellular vesicles, and used Nanoparticle Tracking Analysis to quantify exosomes following implementation of these steps.
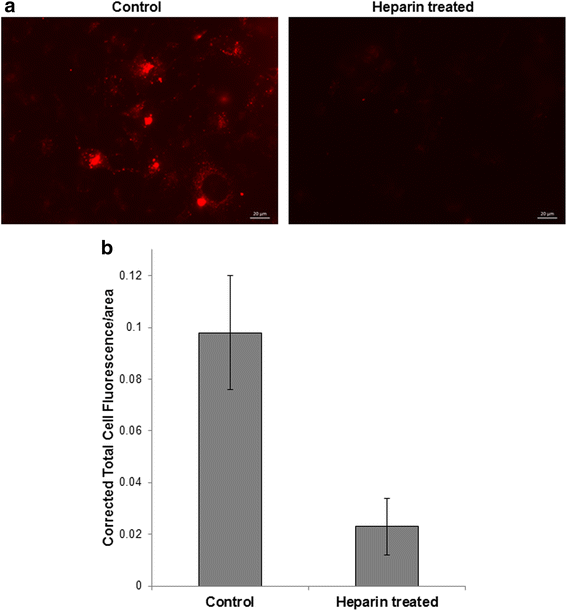
Results: The experimental approach outlined here effectively reduced exosome trafficking by >95%; further use of heparin to block exosome uptake by putative recipient cells further impeded transfer of these extracellular vesicles.
Conclusions: This validated assay incorporates several steps that can be taken to quantifiably control for extracellular vesicles in order to perform studies focused on TNT-selective communication.
Keywords: Exosomes; Extracellular vesicles; Intercellular communication; Intercellular transfer; Membrane nanotubes; Microvesicles; Transwell assay; Tunneling nanotubes.
Conflict of interest statement
Consent for publication
All authors have reviewed the final manuscript prior to submission and consented to its publication.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

