Identifying predictive features of Clostridium difficile infection recurrence before, during, and after primary antibiotic treatment
- PMID: 29132405
- PMCID: PMC5684761
- DOI: 10.1186/s40168-017-0368-1
Identifying predictive features of Clostridium difficile infection recurrence before, during, and after primary antibiotic treatment
Abstract
Background: Colonization by the pathogen Clostridium difficile often occurs in the background of a disrupted microbial community. Identifying specific organisms conferring resistance to invasion by C. difficile is desirable because diagnostic and therapeutic strategies based on the human microbiota have the potential to provide more precision to the management and treatment of Clostridium difficile infection (CDI) and its recurrence.
Methods: We conducted a longitudinal study of adult patients diagnosed with their first CDI. We investigated the dynamics of the gut microbiota during antibiotic treatment, and we used microbial or demographic features at the time of diagnosis, or after treatment, to predict CDI recurrence. To check the validity of the predictions, a meta-analysis using a previously published dataset was performed.
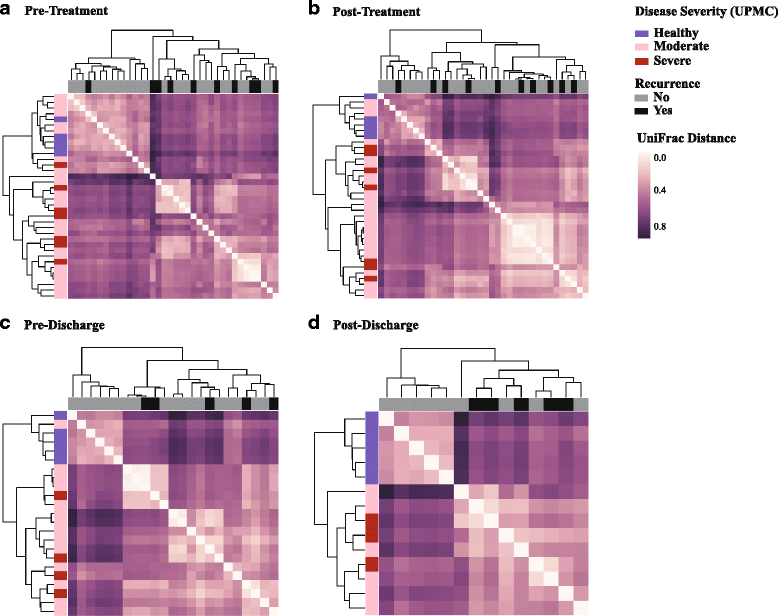
Results: We observed that patients' microbiota "before" antibiotic treatment was predictive of disease relapse, but surprisingly, post-antibiotic microbial community is indistinguishable between patients that recur or not. At the individual OTU level, we identified Veillonella dispar as a candidate organism for preventing CDI recurrence; however, we did not detect a corresponding signal in the conducted meta-analysis.
Conclusion: Although in our patient population, a candidate organism was identified for negatively predicting CDI recurrence, results suggest the need for larger cohort studies that include patients with diverse demographic characteristics to generalize species that robustly confer colonization resistance against C. difficile and accurately predict disease relapse.
Conflict of interest statement
Ethics approval and consent to participate
Written informed consent was obtained from participants at enrollment. This study was approved by the Institutional Review Board (IRB) at State University of New York Downstate Medical Center and the Massachusetts Institute of Technology.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
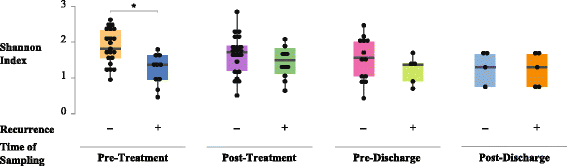



References
-
- Kuijper EJ, Coignard B, Tull P, ESCMID Study Group for Clostridium difficile. European Centre for Disease Prevention and Control Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12:2–18. doi: 10.1111/j.1469-0691.2006.01580.x. - DOI - PubMed
-
- Gravel D, Miller M, Simor A, Taylor G, Gardam M, McGeer A, Hutchinson J, Moore D, Kelly S, Boyd D, et al. Health care-associated Clostridium difficile infection in adults admitted to acute care hospitals in Canada: a Canadian nosocomial infection surveillance program study. Clin Infect Dis. 2009;48:568–576. doi: 10.1086/596703. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

