Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage
- PMID: 29133310
- PMCID: PMC5741051
- DOI: 10.1101/gr.223727.117
Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage
Abstract
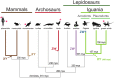
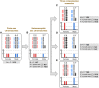
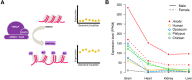
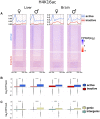
Sex chromosomes differentiated from different ancestral autosomes in various vertebrate lineages. Here, we trace the functional evolution of the XY Chromosomes of the green anole lizard (Anolis carolinensis), on the basis of extensive high-throughput genome, transcriptome and histone modification sequencing data and revisit dosage compensation evolution in representative mammals and birds with substantial new expression data. Our analyses show that Anolis sex chromosomes represent an ancient XY system that originated at least ≈160 million years ago in the ancestor of Iguania lizards, shortly after the separation from the snake lineage. The age of this system approximately coincides with the ages of the avian and two mammalian sex chromosomes systems. To compensate for the almost complete Y Chromosome degeneration, X-linked genes have become twofold up-regulated, restoring ancestral expression levels. The highly efficient dosage compensation mechanism of Anolis represents the only vertebrate case identified so far to fully support Ohno's original dosage compensation hypothesis. Further analyses reveal that X up-regulation occurs only in males and is mediated by a male-specific chromatin machinery that leads to global hyperacetylation of histone H4 at lysine 16 specifically on the X Chromosome. The green anole dosage compensation mechanism is highly reminiscent of that of the fruit fly, Drosophila melanogaster Altogether, our work unveils the convergent emergence of a Drosophila-like dosage compensation mechanism in an ancient reptilian sex chromosome system and highlights that the evolutionary pressures imposed by sex chromosome dosage reductions in different amniotes were resolved in fundamentally different ways.
© 2017 Marin et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
