Mutations in Gene fusA1 as a Novel Mechanism of Aminoglycoside Resistance in Clinical Strains of Pseudomonas aeruginosa
- PMID: 29133559
- PMCID: PMC5786765
- DOI: 10.1128/AAC.01835-17
Mutations in Gene fusA1 as a Novel Mechanism of Aminoglycoside Resistance in Clinical Strains of Pseudomonas aeruginosa
Abstract
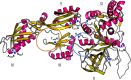
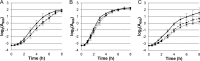
Resistance of clinical strains of Pseudomonas aeruginosa to aminoglycosides can result from production of transferable aminoglycoside-modifying enzymes, of 16S rRNA methylases, and/or mutational derepression of intrinsic multidrug efflux pump MexXY(OprM). We report here the characterization of a new type of mutant that is 4- to 8-fold more resistant to 2-deoxystreptamine derivatives (e.g., gentamicin, amikacin, and tobramycin) than the wild-type strain PAO1. The genetic alterations of three in vitro mutants were mapped on fusA1 and found to result in single amino acid substitutions in domains II, III, and V of elongation factor G (EF-G1A), a key component of translational machinery. Transfer of the mutated fusA1 alleles into PAO1 reproduced the resistance phenotype. Interestingly, fusA1 mutants with other amino acid changes in domains G, IV, and V of EF-G1A were identified among clinical strains with decreased susceptibility to aminoglycosides. Allelic-exchange experiments confirmed the relevance of these latter mutations and of three other previously reported alterations located in domains G and IV. Pump MexXY(OprM) partly contributed to the resistance conferred by the mutated EF-G1A variants and had additive effects on aminoglycoside MICs when mutationally upregulated. Altogether, our data demonstrate that cystic fibrosis (CF) and non-CF strains of P. aeruginosa can acquire a therapeutically significant resistance to important aminoglycosides via a new mechanism involving mutations in elongation factor EF-G1A.
Keywords: EF-G1A; Pseudomonas aeruginosa; aminoglycosides; mechanisms of resistance.
Copyright © 2018 American Society for Microbiology.
Figures
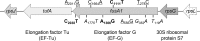
References
-
- Cabot G, Ocampo-Sosa AA, Dominguez MA, Gago JF, Juan C, Tubau F, Rodriguez C, Moya B, Pena C, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious Diseases. 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. - DOI - PMC - PubMed
-
- Shortridge D, Castanheira M, Pfaller MA, Flamm RK. 2017. Ceftolozane-tazobactam activity against Pseudomonas aeruginosa clinical isolates from U.S. Hospitals: report from the PACTS Antimicrobial Surveillance Program, 2012 to 2015. Antimicrob Agents Chemother 61:e00465-17. doi: 10.1128/AAC.00465-17. - DOI - PMC - PubMed
-
- Davies J, Davis BD. 1968. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics: the effect of drug concentration. J Biol Chem 243:3312–3316. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

