The Mutational Landscape of Gastrointestinal Malignancies as Reflected by Circulating Tumor DNA
- PMID: 29133621
- PMCID: PMC5752585
- DOI: 10.1158/1535-7163.MCT-17-0360
The Mutational Landscape of Gastrointestinal Malignancies as Reflected by Circulating Tumor DNA
Abstract
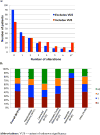
We aimed to assess the utility of a novel, noninvasive method of detecting genomic alterations in patients with gastrointestinal malignancies, i.e., the use of liquid biopsies to obtain blood-derived circulating tumor DNA (ctDNA) through an analysis of the genomic landscape of ctDNA (68 genes) from 213 patients with advanced gastrointestinal cancers. The most common cancer types were colorectal adenocarcinoma (N = 55; 26%), appendiceal adenocarcinoma (N = 46; 22%), hepatocellular carcinoma (N = 31; 15%), and pancreatic ductal adenocarcinoma (N = 25; 12%). The majority of patients (58%) had ≥1 characterized alteration (excluded variants of unknown significance). The median number of characterized alterations was 1 (range, 0-13). The number of detected alterations per patient varied between different cancer types: in hepatocellular carcinoma, 74% of patients (23/31) had ≥1 characterized alteration(s) versus 24% of appendiceal adenocarcinoma patients (11/46). The median percent ctDNA among characterized alterations was 2.50% (interquartile range, 0.76%-8.96%). Overall, 95% of patients (117/123) had distinct molecular portfolios with 143 unique characterized alterations within 56 genes. Overall, concordance rates of 96%, 94%, 95%, and 91%, respectively, were found between ctDNA and tissue biopsy (N = 105 patients) in the four most common alterations (KRAS amplification, MYC amplification, KRAS G12V, and EGFR amplification). Of 123 patients with characterized alterations, >99% (122/123; 57% of entire population tested; 122/213) had one or more alterations potentially actionable by experimental or approved drugs. These observations suggest that many patients with gastrointestinal tumors, including difficult-to-biopsy malignancies like hepatocellular cancers, frequently have discernible and theoretically pharmacologically tractable ctDNA alterations that merit further studies in prospective trials. Mol Cancer Ther; 17(1); 297-305. ©2017 AACR.
©2017 American Association for Cancer Research.
Figures


Similar articles
-
Correlation of Somatic Genomic Alterations Between Tissue Genomics and ctDNA Employing Next-Generation Sequencing: Analysis of Lung and Gastrointestinal Cancers.Mol Cancer Ther. 2018 May;17(5):1123-1132. doi: 10.1158/1535-7163.MCT-17-1015. Epub 2018 Mar 2. Mol Cancer Ther. 2018. PMID: 29500272
-
Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer.J Hematol Oncol. 2019 Dec 4;12(1):130. doi: 10.1186/s13045-019-0824-4. J Hematol Oncol. 2019. PMID: 31801585 Free PMC article.
-
Hybrid Capture-Based Genomic Profiling of Circulating Tumor DNA from Patients with Advanced Cancers of the Gastrointestinal Tract or Anus.Clin Cancer Res. 2018 Apr 15;24(8):1881-1890. doi: 10.1158/1078-0432.CCR-17-3103. Epub 2018 Jan 23. Clin Cancer Res. 2018. PMID: 29363525 Free PMC article.
-
Clinical Utility of Analyzing Circulating Tumor DNA in Patients with Metastatic Colorectal Cancer.Oncologist. 2018 Nov;23(11):1310-1318. doi: 10.1634/theoncologist.2017-0621. Epub 2018 Apr 26. Oncologist. 2018. PMID: 29700206 Free PMC article. Review.
-
Development of circulating tumour DNA analysis for gastrointestinal cancers.ESMO Open. 2020 Jan;5(Suppl 1):e000600. doi: 10.1136/esmoopen-2019-000600. ESMO Open. 2020. PMID: 32830648 Free PMC article. Review.
Cited by
-
Assessing the Concordance of Genomic Alterations between Circulating-Free DNA and Tumour Tissue in Cancer Patients.Cancers (Basel). 2019 Dec 4;11(12):1938. doi: 10.3390/cancers11121938. Cancers (Basel). 2019. PMID: 31817150 Free PMC article. Review.
-
Feasibility and clinical value of circulating tumor DNA testing in patients with gastric adenocarcinomas.J Gastrointest Oncol. 2019 Jun;10(3):400-406. doi: 10.21037/jgo.2019.01.14. J Gastrointest Oncol. 2019. PMID: 31183188 Free PMC article.
-
Prognostic Utility of Pre- and Postoperative Circulating Tumor DNA Liquid Biopsies in Patients with Peritoneal Metastases.Ann Surg Oncol. 2020 Sep;27(9):3259-3267. doi: 10.1245/s10434-020-08331-x. Epub 2020 Aug 6. Ann Surg Oncol. 2020. PMID: 32767050 Free PMC article.
-
Evaluating Pancreatic and Biliary Neoplasms with Small Biopsy-Based Next Generation Sequencing (NGS): Doing More with Less.Cancers (Basel). 2022 Jan 13;14(2):397. doi: 10.3390/cancers14020397. Cancers (Basel). 2022. PMID: 35053560 Free PMC article. Review.
-
The Current Role of Circulating Tumor DNA in the Management of Pancreatic Cancer.J Gastrointest Cancer. 2025 Jan 14;56(1):44. doi: 10.1007/s12029-024-01129-0. J Gastrointest Cancer. 2025. PMID: 39808248 Review.
References
-
- Wheler JJ, Janku F, Naing A, Li Y, Stephen B, Zinner R, et al. Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer Res [Internet] 2016 76:3690–701. Available from: http://cancerres.aacrjournals.org/cgi/doi/10.1158/0008-5472.CAN-15-3043. - DOI - PubMed
-
- Tsimberidou A-M, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res [Internet] Clinical Cancer Research. 2012 [cited 2016 Aug 11];18:6373–83. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22966018. - PMC - PubMed
-
- Von Hoff DD, Stephenson JJ, Rosen P, Loesch DM, Borad MJ, Anthony S, et al. J Clin Oncol [Internet] American Society of Clinical Oncology; 2010. Pilot Study Using Molecular Profiling of Patients’ Tumors to Find Potential Targets and Select Treatments for Their Refractory Cancers. [cited 2016 Aug 11];28:4877–83. Available from: http://jco.ascopubs.org/cgi/doi/10.1200/JCO.2009.26.5983. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

