Protein-Ligand Blind Docking Using QuickVina-W With Inter-Process Spatio-Temporal Integration
- PMID: 29133831
- PMCID: PMC5684369
- DOI: 10.1038/s41598-017-15571-7
Protein-Ligand Blind Docking Using QuickVina-W With Inter-Process Spatio-Temporal Integration
Abstract
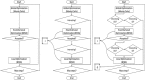
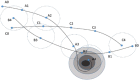
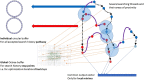
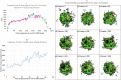
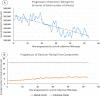
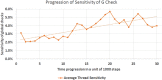
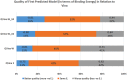
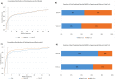
"Virtual Screening" is a common step of in silico drug design, where researchers screen a large library of small molecules (ligands) for interesting hits, in a process known as "Docking". However, docking is a computationally intensive and time-consuming process, usually restricted to small size binding sites (pockets) and small number of interacting residues. When the target site is not known (blind docking), researchers split the docking box into multiple boxes, or repeat the search several times using different seeds, and then merge the results manually. Otherwise, the search time becomes impractically long. In this research, we studied the relation between the search progression and Average Sum of Proximity relative Frequencies (ASoF) of searching threads, which is closely related to the search speed and accuracy. A new inter-process spatio-temporal integration method is employed in Quick Vina 2, resulting in a new docking tool, QuickVina-W, a suitable tool for "blind docking", (not limited in search space size or number of residues). QuickVina-W is faster than Quick Vina 2, yet better than AutoDock Vina. It should allow researchers to screen huge ligand libraries virtually, in practically short time and with high accuracy without the need to define a target pocket beforehand.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Walters WP, Stahl MT, Murcko MA. Virtual screening - an overview. Drug Discovery Today. 1998;3:160–178. doi: 10.1016/S1359-6446(97)01163-X. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

