Network inference from glycoproteomics data reveals new reactions in the IgG glycosylation pathway
- PMID: 29133956
- PMCID: PMC5684356
- DOI: 10.1038/s41467-017-01525-0
Network inference from glycoproteomics data reveals new reactions in the IgG glycosylation pathway
Erratum in
-
Publisher Correction: Network inference from glycoproteomics data reveals new reactions in the IgG glycosylation pathway.Nat Commun. 2018 Feb 13;9(1):706. doi: 10.1038/s41467-017-02379-2. Nat Commun. 2018. PMID: 29440641 Free PMC article.
Abstract
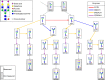
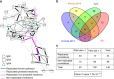
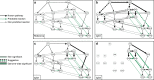
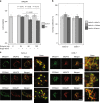
Immunoglobulin G (IgG) is a major effector molecule of the human immune response, and aberrations in IgG glycosylation are linked to various diseases. However, the molecular mechanisms underlying protein glycosylation are still poorly understood. We present a data-driven approach to infer reactions in the IgG glycosylation pathway using large-scale mass-spectrometry measurements. Gaussian graphical models are used to construct association networks from four cohorts. We find that glycan pairs with high partial correlations represent enzymatic reactions in the known glycosylation pathway, and then predict new biochemical reactions using a rule-based approach. Validation is performed using data from a GWAS and results from three in vitro experiments. We show that one predicted reaction is enzymatically feasible and that one rejected reaction does not occur in vitro. Moreover, in contrast to previous knowledge, enzymes involved in our predictions colocalize in the Golgi of two cell lines, further confirming the in silico predictions.
Conflict of interest statement
G.L. declares that he is a founder and owner of Genos, a private research organization that specializes in high-throughput glycomics and has several patents in the field. M.P.-B., I.T.-A., G.R., J.S., L.K. and M.V. are employees of Genos. The remaining authors declare no competing financial interests.
Figures




References
-
- Lauc G, et al. Glycans—the third revolution in evolution. Biochim. Biophys. Acta. 2014;5:4736–4739.
-
- Gerald BP, Jeffrey BL, Lee MW. Immunology, Infection, and Immunity. Washington DC: ASM Press; 2004.
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

