Aligning evidence: concerns regarding multiple sequence alignments in estimating the phylogeny of the Nudibranchia suborder Doridina
- PMID: 29134101
- PMCID: PMC5666284
- DOI: 10.1098/rsos.171095
Aligning evidence: concerns regarding multiple sequence alignments in estimating the phylogeny of the Nudibranchia suborder Doridina
Abstract
Molecular estimates of phylogenetic relationships rely heavily on multiple sequence alignment construction. There has been little consensus, however, on how to properly address issues pertaining to the alignment of variable regions. Here, we construct alignments from four commonly sequenced molecular markers (16S, 18S, 28S and cytochrome c oxidase subunit I) for the Nudibranchia using three different methodologies: (i) strict mathematical algorithm; (ii) exclusion of variable or divergent regions and (iii) manually curated, and examine how different alignment construction methods can affect phylogenetic signal and phylogenetic estimates for the suborder Doridina. Phylogenetic informativeness (PI) profiles suggest that the molecular markers tested lack the power to resolve relationships at the base of the Doridina, while being more robust at family-level classifications. This supports the lack of consistent resolution between the 19 families within the Doridina across all three alignments. Most of the 19 families were recovered as monophyletic, and instances of non-monophyletic families were consistently recovered between analyses. We conclude that the alignment of variable regions has some effect on phylogenetic estimates of the Doridina, but these effects can vary depending on the size and scope of the phylogenetic query and PI of molecular markers.
Keywords: Cryptobranchia; Dorid systematics; Phanerobranchia; multiple sequence alignment; sequence homology.
Conflict of interest statement
We declare that we have no competing interests.
Figures



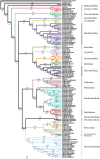
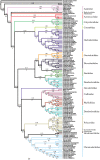
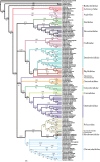
References
-
- Morrison DA, Morgan MJ, Kelchner SA. 2015. Molecular homology and multiple-sequence alignment: an analysis of concepts and practice. Aust. Syst. Bot. 28, 46–62. (doi:10.1071/SB15001) - DOI
-
- Morgan MJ, Kelchner SA. 2010. Inference of molecular homology and sequence alignment by direct optimization. Mol. Phylogenet. Evol. 56, 305–311. (doi:10.1016/j.ympev.2010.03.032) - DOI - PubMed
-
- Morrison DA. 2015. Is sequence alignment an art or a science? Syst. Bot. 40, 14–26. (doi:10.1600/036364415X686305) - DOI
-
- Patterson C. 1988. Homology in classical and molecular biology. Mol. Biol. Evol. 5, 603–625. - PubMed
-
- Smith TF, Waterman MS. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197. (doi:10.1016/0022-2836(81)90087-5) - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
