Transcriptome dynamics along axolotl regenerative development are consistent with an extensive reduction in gene expression heterogeneity in dedifferentiated cells
- PMID: 29134148
- PMCID: PMC5678507
- DOI: 10.7717/peerj.4004
Transcriptome dynamics along axolotl regenerative development are consistent with an extensive reduction in gene expression heterogeneity in dedifferentiated cells
Abstract
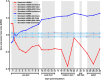
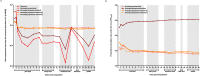
Although in recent years the study of gene expression variation in the absence of genetic or environmental cues or gene expression heterogeneity has intensified considerably, many basic and applied biological fields still remain unaware of how useful the study of gene expression heterogeneity patterns might be for the characterization of biological systems and/or processes. Largely based on the modulator effect chromatin compaction has for gene expression heterogeneity and the extensive changes in chromatin compaction known to occur for specialized cells that are naturally or artificially induced to revert to less specialized states or dedifferentiate, I recently hypothesized that processes that concur with cell dedifferentiation would show an extensive reduction in gene expression heterogeneity. The confirmation of the existence of such trend could be of wide interest because of the biomedical and biotechnological relevance of cell dedifferentiation-based processes, i.e., regenerative development, cancer, human induced pluripotent stem cells, or plant somatic embryogenesis. Here, I report the first empirical evidence consistent with the existence of an extensive reduction in gene expression heterogeneity for processes that concur with cell dedifferentiation by analyzing transcriptome dynamics along forearm regenerative development in Ambystoma mexicanum or axolotl. Also, I briefly discuss on the utility of the study of gene expression heterogeneity dynamics might have for the characterization of cell dedifferentiation-based processes, and the engineering of tools that afforded better monitoring and modulating such processes. Finally, I reflect on how a transitional reduction in gene expression heterogeneity for dedifferentiated cells can promote a long-term increase in phenotypic heterogeneity following cell dedifferentiation with potential adverse effects for biomedical and biotechnological applications.
Keywords: Cell dedifferentiation; Chromatin compaction; Gene expression heterogeneity; Regenerative development.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Somatic cell dedifferentiation/reprogramming for regenerative medicine.Int J Stem Cells. 2009 May;2(1):18-27. doi: 10.15283/ijsc.2009.2.1.18. Int J Stem Cells. 2009. PMID: 24855516 Free PMC article. Review.
-
Preclinical Molecular Signatures of Spinal Cord Functional Restoration: Optimizing the Metamorphic Axolotl (Ambystoma mexicanum) Model in Regenerative Medicine.OMICS. 2020 Jun;24(6):370-378. doi: 10.1089/omi.2020.0024. OMICS. 2020. PMID: 32496969
-
An Identification and Characterization of the Axolotl (Ambystoma mexicanum, Amex) Telomerase Reverse Transcriptase (Amex TERT).Genes (Basel). 2022 Feb 18;13(2):373. doi: 10.3390/genes13020373. Genes (Basel). 2022. PMID: 35205418 Free PMC article.
-
Analysis of embryonic development in the unsequenced axolotl: Waves of transcriptomic upheaval and stability.Dev Biol. 2017 Jun 15;426(2):143-154. doi: 10.1016/j.ydbio.2016.05.024. Epub 2016 Jul 28. Dev Biol. 2017. PMID: 27475628 Free PMC article.
-
Dedifferentiated fat cells: A cell source for regenerative medicine.World J Stem Cells. 2015 Nov 26;7(10):1202-14. doi: 10.4252/wjsc.v7.i10.1202. World J Stem Cells. 2015. PMID: 26640620 Free PMC article. Review.
Cited by
-
Transgenerational Self-Reconstruction of Disrupted Chromatin Organization After Exposure To An Environmental Stressor in Mice.Sci Rep. 2019 Sep 10;9(1):13057. doi: 10.1038/s41598-019-49440-2. Sci Rep. 2019. PMID: 31506492 Free PMC article.
-
Timing Does Matter: Nerve-Mediated HDAC1 Paces the Temporal Expression of Morphogenic Genes During Axolotl Limb Regeneration.Front Cell Dev Biol. 2021 May 10;9:641987. doi: 10.3389/fcell.2021.641987. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34041236 Free PMC article.
References
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Research. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

