ω-3 polyunsaturated fatty acids direct differentiation of the membrane phenotype in mesenchymal stem cells to potentiate osteogenesis
- PMID: 29134198
- PMCID: PMC5677358
- DOI: 10.1126/sciadv.aao1193
ω-3 polyunsaturated fatty acids direct differentiation of the membrane phenotype in mesenchymal stem cells to potentiate osteogenesis
Abstract
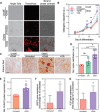
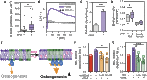
Mammalian cells produce hundreds of dynamically regulated lipid species that are actively turned over and trafficked to produce functional membranes. These lipid repertoires are susceptible to perturbations from dietary sources, with potentially profound physiological consequences. However, neither the lipid repertoires of various cellular membranes, their modulation by dietary fats, nor their effects on cellular phenotypes have been widely explored. We report that differentiation of human mesenchymal stem cells (MSCs) into osteoblasts or adipocytes results in extensive remodeling of the plasma membrane (PM), producing cell-specific membrane compositions and biophysical properties. The distinct features of osteoblast PMs enabled rational engineering of membrane phenotypes to modulate differentiation in MSCs. Specifically, supplementation with docosahexaenoic acid (DHA), a lipid component characteristic of osteoblast membranes, induced broad lipidomic remodeling in MSCs that reproduced compositional and structural aspects of the osteoblastic PM phenotype. The PM changes induced by DHA supplementation potentiated osteogenic differentiation of MSCs concurrent with enhanced Akt activation at the PM. These observations prompt a model wherein the DHA-induced lipidome leads to more stable membrane microdomains, which serve to increase Akt activity and thereby enhance osteogenic differentiation. More broadly, our investigations suggest a general mechanism by which dietary fats affect cellular physiology through remodeling of membrane lipidomes, biophysical properties, and signaling.
Figures



References
-
- Gerl M. J., Sampaio J. L., Urban S., Kalvodova L., Verbavatz J.-M., Binnington B., Lindemann D., Lingwood C. A., Shevchenko A., Schroeder C., Simons K., Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 196, 213–221 (2012). - PMC - PubMed
-
- Klemm R. W., Ejsing C. S., Surma M. A., Kaiser H.-J., Gerl M. J., Sampaio J. L., de Robillard Q., Ferguson C., Proszynski T. J., Shevchenko A., Simons K., Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 185, 601–612 (2009). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

