Artificial intelligence in neurodegenerative disease research: use of IBM Watson to identify additional RNA-binding proteins altered in amyotrophic lateral sclerosis
- PMID: 29134320
- PMCID: PMC5773659
- DOI: 10.1007/s00401-017-1785-8
Artificial intelligence in neurodegenerative disease research: use of IBM Watson to identify additional RNA-binding proteins altered in amyotrophic lateral sclerosis
Abstract
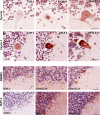
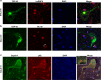
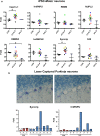
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease with no effective treatments. Numerous RNA-binding proteins (RBPs) have been shown to be altered in ALS, with mutations in 11 RBPs causing familial forms of the disease, and 6 more RBPs showing abnormal expression/distribution in ALS albeit without any known mutations. RBP dysregulation is widely accepted as a contributing factor in ALS pathobiology. There are at least 1542 RBPs in the human genome; therefore, other unidentified RBPs may also be linked to the pathogenesis of ALS. We used IBM Watson® to sieve through all RBPs in the genome and identify new RBPs linked to ALS (ALS-RBPs). IBM Watson extracted features from published literature to create semantic similarities and identify new connections between entities of interest. IBM Watson analyzed all published abstracts of previously known ALS-RBPs, and applied that text-based knowledge to all RBPs in the genome, ranking them by semantic similarity to the known set. We then validated the Watson top-ten-ranked RBPs at the protein and RNA levels in tissues from ALS and non-neurological disease controls, as well as in patient-derived induced pluripotent stem cells. 5 RBPs previously unlinked to ALS, hnRNPU, Syncrip, RBMS3, Caprin-1 and NUPL2, showed significant alterations in ALS compared to controls. Overall, we successfully used IBM Watson to help identify additional RBPs altered in ALS, highlighting the use of artificial intelligence tools to accelerate scientific discovery in ALS and possibly other complex neurological disorders.
Keywords: Amyotrophic lateral sclerosis; Artificial intelligence; Motor neuron; Protein aggregation; RNA-binding protein.
Conflict of interest statement
RB is a founder of Iron Horse Diagnostics, Inc., a biotechnology company focused on diagnostic and prognostic biomarkers for ALS and other neurologic disorders.
Figures





References
-
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. - DOI - PubMed
-
- Bain JM, Cho MT, Telegrafi A, Wilson A, Brooks S, Botti C, Gowans G, Autullo LA, Krishnamurthy V, Willing MC, Toler TL, Ben-Zev B, Elpeleg O, Shen Y, Retterer K, Monaghan KG, Chung WK. Variants in HNRNPH2 on the X chromosome are associated with a neurodevelopmental disorder in females. Am J Hum Genet. 2016;99:728–734. doi: 10.1016/j.ajhg.2016.06.028. - DOI - PMC - PubMed
-
- Blokhuis AM, Koppers M, Groen EJ, van den Heuvel DM, Dini Modigliani S, Anink JJ, Fumoto K, van Diggelen F, Snelting A, Sodaar P, Verheijen BM, Demmers JA, Veldink JH, Aronica E, Bozzoni I, den Hertog J, van den Berg LH, Pasterkamp RJ. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 2016;132:175–196. doi: 10.1007/s00401-016-1575-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

