Soybean mosaic virus: a successful potyvirus with a wide distribution but restricted natural host range
- PMID: 29134790
- PMCID: PMC6638002
- DOI: 10.1111/mpp.12644
Soybean mosaic virus: a successful potyvirus with a wide distribution but restricted natural host range
Abstract
Taxonomy: Soybean mosaic virus (SMV) is a species within the genus Potyvirus, family Potyviridae, which includes almost one-quarter of all known plant RNA viruses affecting agriculturally important plants. The Potyvirus genus is the largest of all genera of plant RNA viruses with 160 species.
Particle: The filamentous particles of SMV, typical of potyviruses, are about 7500 Å long and 120 Å in diameter with a central hole of about 15 Å in diameter. Coat protein residues are arranged in helices of about 34 Å pitch having slightly less than nine subunits per turn.
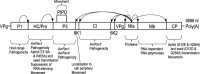
Genome: The SMV genome consists of a single-stranded, positive-sense, polyadenylated RNA of approximately 9.6 kb with a virus-encoded protein (VPg) linked at the 5' terminus. The genomic RNA contains a single large open reading frame (ORF). The polypeptide produced from the large ORF is processed proteolytically by three viral-encoded proteinases to yield about 10 functional proteins. A small ORF, partially overlapping the P3 cistron, pipo, is encoded as a fusion protein in the N-terminus of P3 (P3N + PIPO).
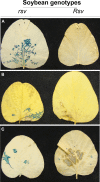
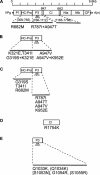
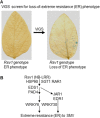
Biological properties: SMV's host range is restricted mostly to two plant species of a single genus: Glycine max (cultivated soybean) and G. soja (wild soybean). SMV is transmitted by aphids non-persistently and by seeds. The variability of SMV is recognized by reactions on cultivars with dominant resistance (R) genes. Recessive resistance genes are not known.
Geographical distribution and economic importance: As a consequence of its seed transmissibility, SMV is present in all soybean-growing areas of the world. SMV infections can reduce significantly seed quantity and quality (e.g. mottled seed coats, reduced seed size and viability, and altered chemical composition).
Control: The most effective means of managing losses from SMV are the planting of virus-free seeds and cultivars containing single or multiple R genes.
Key attractions: The interactions of SMV with soybean genotypes containing different dominant R genes and an understanding of the functional role(s) of SMV-encoded proteins in virulence, transmission and pathogenicity have been investigated intensively. The SMV-soybean pathosystem has become an excellent model for the examination of the genetics and genomics of a uniquely complex gene-for-gene resistance model in a crop of worldwide importance.
Keywords: Potyviridae; R genes; RNA viruses; avirulence/virulence proteins; host responses; signalling; virus-host interactions.
© 2017 BSPP AND JOHN WILEY & SONS LTD.
Figures

References
-
- Adams, M.J. , Zerbini, F.M. , French, R. , Rabenstein, F. , Stenger, D.C. and Valkonen, J.P.T. (2012). Potyviridae In: Virus Taxonomy . Ninth Report of the International Committee on Taxonomy of Viruses (King A.M.Q., Adams M.J., Carstens E.B. and Lefkowitz E.J., eds)., pp. 1069–1089. London: Elsevier/Academic Press.
-
- Ahangaran, A. , Habibi, M.K. , Mohammadi, G.‐H.M. , Winter, S. and Garcia‐Arenal, F. (2013) Analysis of Soybean mosaic virus genetic diversity in Iran allows the characterization of a new mutation resulting in overcoming Rsv4‐resistance. J. Gen. Virol. 94, 2557–2568. - PubMed
-
- Almeida, A.M.R. (1981) Identification of strains of soybean common mosaic virus in Parana State. Fitopatol. Bras. 6, 131–136.
-
- Almeida, A.M.R. , Sakai, J. , Souto, E.R. , Kitajima, E.W. , Fukuji, T.S. and Hanada, K. (2002) Mosaic in Senna occidentalis in southern Brazil induced by a new strain of Soybean mosaic virus . Fitopatol. Bras. 27, 151–156.
-
- Anjos, J.R.M. , Lin, M.T. and Kitajima, E.W. (1985) Characterization of an isolate of soybean mosaic virus. Fitopatol. Bras. 10, 143–157.
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

