Generation and testing anti-influenza human monoclonal antibodies in a new humanized mouse model (DRAGA: HLA-A2. HLA-DR4. Rag1 KO. IL-2Rγc KO. NOD)
- PMID: 29135340
- PMCID: PMC5806689
- DOI: 10.1080/21645515.2017.1403703
Generation and testing anti-influenza human monoclonal antibodies in a new humanized mouse model (DRAGA: HLA-A2. HLA-DR4. Rag1 KO. IL-2Rγc KO. NOD)
Abstract
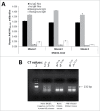
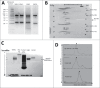
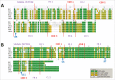
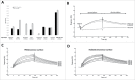
Pandemic outbreaks of influenza type A viruses have resulted in numerous fatalities around the globe. Since the conventional influenza vaccines (CIV) provide less than 20% protection for individuals with weak immune system, it has been considered that broadly cross-neutralizing antibodies may provide a better protection. Herein, we showed that a recently generated humanized mouse (DRAGA mouse; HLA-A2. HLA-DR4. Rag1KO. IL-2Rgc KO. NOD) that lacks the murine immune system and expresses a functional human immune system can be used to generate cross-reactive, human anti-influenza monoclonal antibodies (hu-mAb). DRAGA mouse was also found to be suitable for influenza virus infection, as it can clear a sub-lethal infection and sustain a lethal infection with PR8/A/34 influenza virus. The hu-mAbs were designed for targeting a human B-cell epitope (180WGIHHPPNSKEQ QNLY195) of hemagglutinin (HA) envelope protein of PR8/A/34 (H1N1) virus with high homology among seven influenza type A viruses. A single administration of HA180-195 specific hu-mAb in PR8-infected DRAGA mice significantly delayed the lethality by reducing the lung damage. The results demonstrated that DRAGA mouse is a suitable tool to (i) generate heterotype cross-reactive, anti-influenza human monoclonal antibodies, (ii) serve as a humanized mouse model for influenza infection, and (iii) assess the efficacy of anti-influenza antibody-based therapeutics for human use.
Keywords: Anti-flu antibody therapy; Human influenza infection model; Human monoclonal antibodies; Humanized mice.
Figures




References
-
- Kilbourne ED. Influenza. New York, NY: Plenum Medical Book Co.; 1987
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
