A Role of Sp1 Binding Motifs in Basal and Large T-Antigen-Induced Promoter Activities of Human Polyomavirus HPyV9 and Its Variant UF-1
- PMID: 29135936
- PMCID: PMC5713382
- DOI: 10.3390/ijms18112414
A Role of Sp1 Binding Motifs in Basal and Large T-Antigen-Induced Promoter Activities of Human Polyomavirus HPyV9 and Its Variant UF-1
Abstract
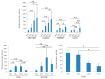
Human polyomavirus 9 (HPyV9) was originally detected in the serum of a renal transplant patient. Seroepidemiological studies showed that ~20-50% of the human population have antibodies against this virus. HPyV9 has not yet been associated with any disease and little is known about the route of infection, transmission, host cell tropism, and genomic variability in circulating strains. Recently, the HPyV9 variant UF-1 with an eight base-pair deletion, a thirteen base-pair insertion and with point mutations, creating three putative Sp1 binding sites in the late promoter was isolated from an AIDS patient. Transient transfection studies with a luciferase reporter plasmid driven by HPyV9 or UF1 promoter demonstrated that UF1 early and late promoters were stronger than HPyV9 promoters in most cell lines, and that the UF1 late promoter was more potently activated by HPyV9 large T-antigen (LTAg). Mutation of two Sp1 motifs strongly reduced trans-activation of the late UF1 promoter by HPyV9 LTAg in HeLa cells. In conclusion, the mutations in the UF1 late promoter seem to strengthen its activity and its response to stimulation by HPyV9 LTAg in certain cells. It remains to be investigated whether these promoter changes have an influence on virus replication and affect the possible pathogenic properties of the virus.
Keywords: Sp1; large T antigen; luciferase; mutation; non-coding control region.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

