WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research
- PMID: 29136241
- PMCID: PMC5753270
- DOI: 10.1093/nar/gkx1064
WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research
Abstract
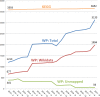
WikiPathways (wikipathways.org) captures the collective knowledge represented in biological pathways. By providing a database in a curated, machine readable way, omics data analysis and visualization is enabled. WikiPathways and other pathway databases are used to analyze experimental data by research groups in many fields. Due to the open and collaborative nature of the WikiPathways platform, our content keeps growing and is getting more accurate, making WikiPathways a reliable and rich pathway database. Previously, however, the focus was primarily on genes and proteins, leaving many metabolites with only limited annotation. Recent curation efforts focused on improving the annotation of metabolism and metabolic pathways by associating unmapped metabolites with database identifiers and providing more detailed interaction knowledge. Here, we report the outcomes of the continued growth and curation efforts, such as a doubling of the number of annotated metabolite nodes in WikiPathways. Furthermore, we introduce an OpenAPI documentation of our web services and the FAIR (Findable, Accessible, Interoperable and Reusable) annotation of resources to increase the interoperability of the knowledge encoded in these pathways and experimental omics data. New search options, monthly downloads, more links to metabolite databases, and new portals make pathway knowledge more effortlessly accessible to individual researchers and research communities.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

