Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer
- PMID: 29136504
- PMCID: PMC5728182
- DOI: 10.1016/j.ccell.2017.10.007
Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer
Erratum in
-
Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer.Cancer Cell. 2018 Jan 8;33(1):151-152. doi: 10.1016/j.ccell.2017.12.011. Cancer Cell. 2018. PMID: 29316428 Free PMC article. No abstract available.
Abstract
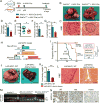
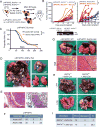
ARID1A, an SWI/SNF chromatin-remodeling gene, is commonly mutated in cancer and hypothesized to be tumor suppressive. In some hepatocellular carcinoma patients, ARID1A was highly expressed in primary tumors but not in metastatic lesions, suggesting that ARID1A can be lost after initiation. Mice with liver-specific homozygous or heterozygous Arid1a loss were resistant to tumor initiation while ARID1A overexpression accelerated initiation. In contrast, homozygous or heterozygous Arid1a loss in established tumors accelerated progression and metastasis. Mechanistically, gain of Arid1a function promoted initiation by increasing CYP450-mediated oxidative stress, while loss of Arid1a within tumors decreased chromatin accessibility and reduced transcription of genes associated with migration, invasion, and metastasis. In summary, ARID1A has context-dependent tumor-suppressive and oncogenic roles in cancer.
Keywords: ARID1A; SWI/SNF chromatin-remodeling complex; epigenetics; hepatocellular carcinoma; metastasis; mouse models.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures






Comment in
-
A Two-Faced mSWI/SNF Subunit: Dual Roles for ARID1A in Tumor Suppression and Oncogenicity in the Liver.Cancer Cell. 2017 Nov 13;32(5):542-543. doi: 10.1016/j.ccell.2017.10.014. Cancer Cell. 2017. PMID: 29136498
References
-
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S-I, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature Genetics. 2012;44:760–764. doi: 10.1038/ng.2291. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

