RLIM suppresses hepatocellular carcinogenesis by up-regulating p15 and p21
- PMID: 29137325
- PMCID: PMC5669951
- DOI: 10.18632/oncotarget.20904
RLIM suppresses hepatocellular carcinogenesis by up-regulating p15 and p21
Abstract
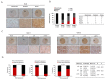
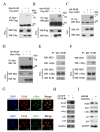
Hepatocellular carcinogenesis results from dysregulation of oncogenes and tumor suppressors that influence cellular proliferation, differentiation and apoptosis. p15 and p21 are cyclin-dependent kinase inhibitors, which arrest cell proliferation and serve as critical tumor suppressors. Here we report that the E3 ubiquitin ligase RLIM expression is downregulated in hepatocellular carcinoma patients, and correlated with p15 and p21 expression in clinical progression. In addition, we showed that RLIM overexpression suppresses the cell growth and arrests cell cycle progression of hepatocellular carcinoma. Mechanistically, we found that RLIM directly binds to MIZ1, disrupting the interaction between c-MYC and MIZ1, and enhancing p15 and p21 transcription. Our results demonstrate that RLIM is an important suppressor in hepatocellular carcinogenesis.
Keywords: MIZ1; RLIM; hepatocellular carcinogenesis; p15; p21.
Conflict of interest statement
CONFLICTS OF INTEREST All authors declare no conflicts of interest.
Figures



Similar articles
-
Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15.Genes Dev. 2013 May 15;27(10):1101-14. doi: 10.1101/gad.214577.113. Genes Dev. 2013. PMID: 23699408 Free PMC article.
-
Cell cycle regulators in bladder cancer: relationship to schistosomiasis.IUBMB Life. 2004 Sep;56(9):557-64. doi: 10.1080/15216540400013903. IUBMB Life. 2004. PMID: 15590562
-
E3 Ubiquitin Ligase RLIM Negatively Regulates c-Myc Transcriptional Activity and Restrains Cell Proliferation.PLoS One. 2016 Sep 29;11(9):e0164086. doi: 10.1371/journal.pone.0164086. eCollection 2016. PLoS One. 2016. PMID: 27684546 Free PMC article.
-
Hippocalcin-like 1 suppresses hepatocellular carcinoma progression by promoting p21(Waf/Cip1) stabilization by activating the ERK1/2-MAPK pathway.Hepatology. 2016 Mar;63(3):880-97. doi: 10.1002/hep.28395. Epub 2016 Feb 3. Hepatology. 2016. PMID: 26659654
-
The expression of P21 and P15 proteins in liver tissue of chronic viral hepatitis and hepatocellular carcinoma and their relation to cell apoptosis.Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2000 Jun;14(2):157-9. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2000. PMID: 11503050
Cited by
-
RNF12 is regulated by AKT phosphorylation and promotes TGF-β driven breast cancer metastasis.Cell Death Dis. 2022 Jan 10;13(1):44. doi: 10.1038/s41419-021-04493-y. Cell Death Dis. 2022. PMID: 35013159 Free PMC article.
-
RNF12 catalyzes BRF1 ubiquitination and regulates RNA polymerase III-dependent transcription.J Biol Chem. 2019 Jan 4;294(1):130-141. doi: 10.1074/jbc.RA118.004524. Epub 2018 Nov 9. J Biol Chem. 2019. PMID: 30413534 Free PMC article.
-
Sequential stabilization of RNF220 by RLIM and ZC4H2 during cerebellum development and Shh-group medulloblastoma progression.J Mol Cell Biol. 2022 Apr 5;14(1):mjab082. doi: 10.1093/jmcb/mjab082. J Mol Cell Biol. 2022. PMID: 35040952 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. https://doi.org/10.3322/caac.20006. - DOI - PubMed
-
- Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–58. https://doi.org/10.1038/nrgastro.2010.100. - DOI - PMC - PubMed
-
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–7. https://doi.org/10.1038/nature03043. - DOI - PubMed
-
- Lin CP, Liu CR, Lee CN, Chan TS, Liu HE. Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J Hepatol. 2010;2:16–20. https://doi.org/10.4254/wjh.v2.i1.16. - DOI - PMC - PubMed
-
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–8. https://doi.org/10.1038/35070086. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

