Trends of long noncoding RNA research from 2007 to 2016: a bibliometric analysis
- PMID: 29137328
- PMCID: PMC5669954
- DOI: 10.18632/oncotarget.20851
Trends of long noncoding RNA research from 2007 to 2016: a bibliometric analysis
Abstract
Purpose: This study aims to analyze the scientific output of long noncoding RNA (lncRNA) research and construct a model to evaluate publications from the past decade qualitatively and quantitatively.
Methods: Publications from 2007 to 2016 were retrieved from the Web of Science Core Collection database. Microsoft Excel 2016 and CiteSpace IV software were used to analyze publication outputs, journals, countries, institutions, authors, citation counts, ESI top papers, H-index, and research frontiers.
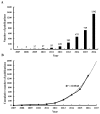
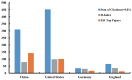
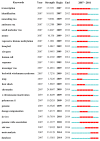
Results: A total of 3,008 papers on lncRNA research were identified published by June 17, 2017. The journal, Oncotarget (IF2016, 5.168) ranked first in the number of publications. China had the largest number of publications (1,843), but the United States showed its dominant position in both citation frequency (45,120) and H-index (97). Zhang Y (72 publications) published the most papers, and Guttman M (1,556 citations) had the greatest co-citation counts. The keyword "database" ranked first in research frontiers.
Conclusion: The annual number of publications rapidly increased in the past decade. China showed its significant progress in lncRNA research, but the United States was the actual leading country in this field. Many Chinese institutions engaged in lncRNA research but significant collaborations among them were not noted. Guttman M, Mercer TR, Rinn JL, and Gupta RA were identified as good candidates for research collaboration. "Database," "Xist RNA," and "Genome-wide association study" should be closely observed in this field.
Keywords: CiteSpace IV; WoSCC; bibliometric; citation; lncRNA.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no financial interests in the findings described in this study.
Figures





References
-
- Perkel JM. Visiting “noncodarnia”. Biotechniques. 2013;54:301, 3–4. https://doi.org/10.2144/000114037. - DOI - PubMed
-
- Morris KV, Mattick JS. The rise of regulatory RNA. Nature reviews Genetics. 2014;15:423–37. https://doi.org/10.1038/nrg3722. - DOI - PMC - PubMed
-
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. https://doi.org/10.1038/nsmb.2942. - DOI - PubMed
-
- Lee JT. Epigenetic Regulation by Long Noncoding RNAs. Science. 2012;338:1435–9. https://doi.org/10.1126/science.1231776. - DOI - PubMed
-
- Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci Rep. 2014;4:6591. https://doi.org/10.1038/srep06591. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

