Genome-wide analysis and functional prediction of long non-coding RNAs in mouse uterus during the implantation window
- PMID: 29137430
- PMCID: PMC5663602
- DOI: 10.18632/oncotarget.21031
Genome-wide analysis and functional prediction of long non-coding RNAs in mouse uterus during the implantation window
Abstract
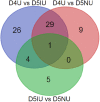
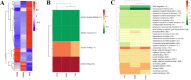
Establishment of the receptive uterus is a crucial step for embryo implantation. In this study, the expression profiles and characterization of long non-coding RNAs (lncRNAs) in pregnant mouse uteri on day 4, day 5 at implantation sites and inter-implantation sites were conducted using RNA-seq. A total of 7,764 putative lncRNA transcripts were identified, including 6,179 known lncRNA transcripts and 1,585 novel lncRNA transcripts. Bioinformatics analysis of the cis and trans lncRNA targets showed that the differentially expressed lncRNAs were mainly involved in tissue remodelling, immune response and metabolism-related processes, indicating that lncRNAs could be involved in the regulation of embryo implantation. We also discovered that differentially expressed lncRNAs might regulate multiple signalling pathways that play an important role in the regulation of embryo implantation. In addition, nine known lncRNAs and four novel lncRNAs were randomly selected and validated by qRT-PCR. The expression of Tug1, Neat1, Gas5, Malat1, H19 and Rmst were significantly regulated in the mouse uterus during the implantation window. Our results are the first to systematically identify lncRNAs in the mouse uterus and provide a catalogue of lncRNAs for further understanding their functions in pregnant mouse uteri during the implantation window.
Keywords: RNA-seq; implantation; long non-coding RNAs; mouse; uterus.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Figures


References
-
- Yoshinaga K. A sequence of events in the uterus prior to implantation in the mouse. J Assist Reprod Genet. 2013;30:1017–22. https://doi.org/10.1007/s10815-013-0093-z. - DOI - PMC - PubMed
-
- Martinez-Conejero JA, Morgan M, Montesinos M, Fortuno S, Meseguer M, Simon C, Horcajadas JA, Pellicer A. Adenomyosis does not affect implantation, but is associated with miscarriage in patients undergoing oocyte donation. Fertil Steril. 2011;96:943–50. https://doi.org/10.1016/j.fertnstert.2011.07.1088. - DOI - PubMed
-
- Cakmak H, Taylor HS. Implantation failure: molecular mechanisms and clinical treatment. Hum Reprod Update. 2011;17:242–53. https://doi.org/10.1093/humupd/dmq037. - DOI - PMC - PubMed
-
- Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24:594–602. https://doi.org/10.1016/j.tcb.2014.06.003. - DOI - PMC - PubMed
-
- Chen G, Shi T, Shi L. Characterizing and annotating the genome using RNA-seq data. Sci China Life Sci. 2017;60:116–25. https://doi.org/10.1007/s11427-015-0349-4. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

